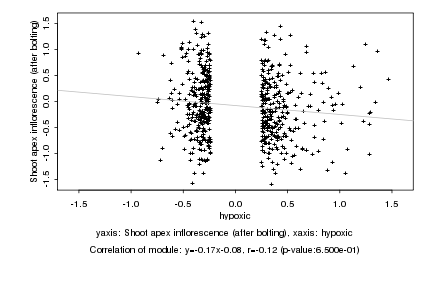
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hypoxic |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.466 |
0.435 |
| 2 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.357 |
0.978 |
| 3 |
247024_at |
AT5G66985
|
unknown |
1.333 |
-0.000 |
| 4 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.291 |
-0.194 |
| 5 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.281 |
-0.430 |
| 6 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.279 |
-0.221 |
| 7 |
250464_at |
AT5G10040
|
unknown |
1.276 |
-1.018 |
| 8 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.241 |
1.099 |
| 9 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.219 |
-0.372 |
| 10 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.198 |
0.280 |
| 11 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.128 |
0.689 |
| 12 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.067 |
-0.905 |
| 13 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.047 |
-1.381 |
| 14 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
1.017 |
-0.057 |
| 15 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
1.007 |
-0.405 |
| 16 |
250152_at |
AT5G15120
|
unknown |
0.987 |
0.159 |
| 17 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.951 |
-0.048 |
| 18 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.931 |
-0.172 |
| 19 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.928 |
-1.173 |
| 20 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.925 |
-0.075 |
| 21 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.919 |
0.083 |
| 22 |
257925_at |
AT3G23170
|
unknown |
0.888 |
0.262 |
| 23 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.874 |
-1.313 |
| 24 |
249384_at |
AT5G39890
|
unknown |
0.862 |
0.569 |
| 25 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.845 |
-0.369 |
| 26 |
261567_at |
AT1G33055
|
unknown |
0.837 |
-0.016 |
| 27 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.837 |
-0.730 |
| 28 |
245226_at |
AT3G29970
|
[B12D protein] |
0.831 |
0.363 |
| 29 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.818 |
0.552 |
| 30 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.791 |
-0.162 |
| 31 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.789 |
-0.957 |
| 32 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.754 |
-0.456 |
| 33 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.752 |
0.381 |
| 34 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.750 |
-0.685 |
| 35 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.747 |
0.555 |
| 36 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.733 |
-1.005 |
| 37 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.716 |
0.141 |
| 38 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.712 |
-0.079 |
| 39 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.691 |
-0.088 |
| 40 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.680 |
-0.936 |
| 41 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.678 |
-0.933 |
| 42 |
248820_at |
AT5G47060
|
unknown |
0.673 |
1.070 |
| 43 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.672 |
0.945 |
| 44 |
254200_at |
AT4G24110
|
unknown |
0.654 |
-0.404 |
| 45 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.648 |
-0.190 |
| 46 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.636 |
-0.920 |
| 47 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.634 |
-0.901 |
| 48 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.632 |
0.165 |
| 49 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.623 |
0.542 |
| 50 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.620 |
-1.296 |
| 51 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.602 |
-0.506 |
| 52 |
252882_at |
AT4G39675
|
unknown |
0.602 |
0.084 |
| 53 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.585 |
-0.859 |
| 54 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.578 |
-0.337 |
| 55 |
261247_at |
AT1G20070
|
unknown |
0.572 |
-0.503 |
| 56 |
260668_at |
AT1G19530
|
unknown |
0.568 |
-0.327 |
| 57 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.566 |
-0.076 |
| 58 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.563 |
-1.035 |
| 59 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.556 |
-0.558 |
| 60 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.543 |
0.231 |
| 61 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.542 |
-1.043 |
| 62 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.536 |
-0.744 |
| 63 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.528 |
-1.176 |
| 64 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
0.527 |
-0.549 |
| 65 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.523 |
0.189 |
| 66 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.521 |
1.273 |
| 67 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.520 |
0.706 |
| 68 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.509 |
-0.514 |
| 69 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.505 |
0.172 |
| 70 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.500 |
0.561 |
| 71 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.494 |
-0.211 |
| 72 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.490 |
-0.228 |
| 73 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.490 |
0.166 |
| 74 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.488 |
-0.089 |
| 75 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.487 |
-0.102 |
| 76 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.485 |
0.334 |
| 77 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.481 |
-0.986 |
| 78 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.481 |
-0.659 |
| 79 |
247488_at |
AT5G61820
|
unknown |
0.479 |
-0.326 |
| 80 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.473 |
-0.662 |
| 81 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.472 |
0.906 |
| 82 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.465 |
-0.064 |
| 83 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.465 |
-1.102 |
| 84 |
256518_at |
AT1G66080
|
unknown |
0.462 |
-0.613 |
| 85 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.461 |
-0.902 |
| 86 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.459 |
0.012 |
| 87 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.459 |
0.045 |
| 88 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.454 |
-0.657 |
| 89 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.454 |
-0.287 |
| 90 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.453 |
-0.111 |
| 91 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.449 |
0.502 |
| 92 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.445 |
-0.439 |
| 93 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.445 |
-0.208 |
| 94 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.442 |
-0.860 |
| 95 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.438 |
-1.000 |
| 96 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.437 |
-0.303 |
| 97 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.437 |
-0.729 |
| 98 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.431 |
0.114 |
| 99 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.430 |
1.204 |
| 100 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.428 |
0.423 |
| 101 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.427 |
-0.499 |
| 102 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.425 |
0.106 |
| 103 |
267470_at |
AT2G30490
|
ATC4H, CINNAMATE 4-HYDROXYLASE, C4H, cinnamate-4-hydroxylase, CYP73A5, REF3, REDUCED EPRDERMAL FLUORESCENCE 3 |
0.424 |
1.451 |
| 104 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.421 |
-0.356 |
| 105 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.421 |
0.206 |
| 106 |
245352_at |
AT4G15490
|
UGT84A3 |
0.419 |
-0.114 |
| 107 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.418 |
0.694 |
| 108 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.415 |
0.723 |
| 109 |
248218_at |
AT5G53710
|
unknown |
0.415 |
-0.033 |
| 110 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.414 |
-0.370 |
| 111 |
253513_at |
AT4G31760
|
[Peroxidase superfamily protein] |
0.410 |
0.485 |
| 112 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.409 |
-0.533 |
| 113 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.409 |
0.160 |
| 114 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.406 |
0.552 |
| 115 |
247324_at |
AT5G64190
|
unknown |
0.406 |
-0.689 |
| 116 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.405 |
-0.296 |
| 117 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.405 |
-0.659 |
| 118 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.403 |
0.489 |
| 119 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.400 |
-0.669 |
| 120 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.399 |
-1.195 |
| 121 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.398 |
-0.733 |
| 122 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.397 |
-0.436 |
| 123 |
256447_at |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
0.396 |
-0.905 |
| 124 |
254069_at |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
0.396 |
-0.709 |
| 125 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.395 |
0.004 |
| 126 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.394 |
-0.701 |
| 127 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.392 |
-0.690 |
| 128 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.392 |
0.326 |
| 129 |
245401_at |
AT4G17670
|
unknown |
0.390 |
0.279 |
| 130 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
0.386 |
0.148 |
| 131 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.386 |
-0.127 |
| 132 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.385 |
-0.869 |
| 133 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.385 |
-0.593 |
| 134 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.384 |
-0.407 |
| 135 |
260005_at |
AT1G67920
|
unknown |
0.380 |
-0.192 |
| 136 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.378 |
-0.130 |
| 137 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.376 |
-0.469 |
| 138 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.375 |
-1.379 |
| 139 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.373 |
-0.653 |
| 140 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.373 |
1.288 |
| 141 |
258493_at |
AT3G02555
|
unknown |
0.370 |
0.371 |
| 142 |
257844_at |
AT3G28480
|
[Oxoglutarate/iron-dependent oxygenase] |
0.368 |
0.078 |
| 143 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
0.368 |
0.129 |
| 144 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.367 |
-0.813 |
| 145 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.365 |
0.272 |
| 146 |
259385_at |
AT1G13470
|
unknown |
0.365 |
-0.671 |
| 147 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.365 |
-0.490 |
| 148 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.365 |
-0.400 |
| 149 |
260327_at |
AT1G63840
|
[RING/U-box superfamily protein] |
0.357 |
-0.027 |
| 150 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.356 |
-1.307 |
| 151 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.355 |
0.681 |
| 152 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.352 |
-1.170 |
| 153 |
260602_at |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
0.352 |
0.446 |
| 154 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.351 |
-0.481 |
| 155 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.350 |
-0.912 |
| 156 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.349 |
0.696 |
| 157 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.348 |
-0.514 |
| 158 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.347 |
-0.179 |
| 159 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.347 |
-0.590 |
| 160 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.346 |
-0.196 |
| 161 |
265539_at |
AT2G15830
|
unknown |
0.344 |
-0.497 |
| 162 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
0.343 |
-1.578 |
| 163 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.342 |
0.687 |
| 164 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.341 |
-0.331 |
| 165 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.340 |
-0.714 |
| 166 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
0.339 |
-0.271 |
| 167 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.337 |
-0.623 |
| 168 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.336 |
0.629 |
| 169 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.332 |
-0.200 |
| 170 |
254263_at |
AT4G23493
|
unknown |
0.330 |
0.290 |
| 171 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.330 |
-0.051 |
| 172 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.329 |
-0.140 |
| 173 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.328 |
-0.561 |
| 174 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.328 |
0.275 |
| 175 |
246125_at |
AT5G19875
|
unknown |
0.328 |
-0.335 |
| 176 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.327 |
-0.238 |
| 177 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.324 |
0.057 |
| 178 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.324 |
-0.457 |
| 179 |
250828_at |
AT5G05250
|
unknown |
0.323 |
-0.805 |
| 180 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.321 |
0.338 |
| 181 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.320 |
-0.997 |
| 182 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.320 |
-0.891 |
| 183 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.319 |
0.427 |
| 184 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.319 |
-0.343 |
| 185 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.319 |
0.050 |
| 186 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.318 |
-0.179 |
| 187 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.318 |
-0.497 |
| 188 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.316 |
-0.795 |
| 189 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.314 |
-0.656 |
| 190 |
257375_at |
AT2G38640
|
unknown |
0.314 |
-1.086 |
| 191 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.313 |
-0.484 |
| 192 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.310 |
0.054 |
| 193 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
0.309 |
1.051 |
| 194 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.308 |
0.613 |
| 195 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.308 |
-0.988 |
| 196 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.308 |
0.550 |
| 197 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.308 |
0.799 |
| 198 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.308 |
-0.656 |
| 199 |
257545_at |
AT3G23200
|
[Uncharacterised protein family (UPF0497)] |
0.308 |
0.055 |
| 200 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.307 |
-0.281 |
| 201 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.307 |
-0.303 |
| 202 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.307 |
-0.493 |
| 203 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.307 |
0.253 |
| 204 |
253393_at |
AT4G32690
|
ATGLB3, ARABIDOPSIS HEMOGLOBIN 3, GLB3, hemoglobin 3 |
0.306 |
-0.556 |
| 205 |
247851_at |
AT5G58070
|
ATTIL, TEMPERATURE-INDUCED LIPOCALIN, TIL, temperature-induced lipocalin |
0.306 |
0.045 |
| 206 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.305 |
-0.512 |
| 207 |
245905_at |
AT5G11090
|
[serine-rich protein-related] |
0.304 |
0.776 |
| 208 |
262751_at |
AT1G16310
|
[Cation efflux family protein] |
0.303 |
-0.141 |
| 209 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
0.301 |
-0.421 |
| 210 |
263250_at |
AT2G31390
|
[pfkB-like carbohydrate kinase family protein] |
0.300 |
0.158 |
| 211 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.299 |
-0.356 |
| 212 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
0.299 |
0.361 |
| 213 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.298 |
0.215 |
| 214 |
262014_at |
AT1G35660
|
unknown |
0.297 |
0.041 |
| 215 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.296 |
-0.805 |
| 216 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
0.295 |
1.335 |
| 217 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.294 |
0.369 |
| 218 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.292 |
-0.404 |
| 219 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.292 |
-0.684 |
| 220 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.290 |
0.026 |
| 221 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.288 |
-0.387 |
| 222 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.288 |
-0.382 |
| 223 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.288 |
0.435 |
| 224 |
264635_at |
AT1G65500
|
unknown |
0.288 |
0.393 |
| 225 |
256262_at |
AT3G12150
|
unknown |
0.287 |
-0.711 |
| 226 |
248706_at |
AT5G48530
|
unknown |
0.285 |
-0.801 |
| 227 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.284 |
0.069 |
| 228 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.284 |
0.077 |
| 229 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.283 |
0.138 |
| 230 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.283 |
-0.451 |
| 231 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.282 |
0.354 |
| 232 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.282 |
-0.028 |
| 233 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.281 |
-0.278 |
| 234 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.281 |
-0.207 |
| 235 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.279 |
1.177 |
| 236 |
262583_at |
AT1G15110
|
PSS1, phosphatidylserine synthase 1 |
0.279 |
0.792 |
| 237 |
266416_at |
AT2G38710
|
[AMMECR1 family] |
0.278 |
0.706 |
| 238 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.277 |
0.784 |
| 239 |
267054_at |
AT2G38370
|
unknown |
0.276 |
1.104 |
| 240 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.274 |
-0.217 |
| 241 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.274 |
-0.503 |
| 242 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.274 |
-0.662 |
| 243 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.274 |
-0.723 |
| 244 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.273 |
1.177 |
| 245 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.273 |
0.027 |
| 246 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.273 |
-0.807 |
| 247 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.272 |
0.034 |
| 248 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.271 |
-0.655 |
| 249 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.271 |
-0.759 |
| 250 |
253965_at |
AT4G26490
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.271 |
-0.975 |
| 251 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.270 |
-0.203 |
| 252 |
264082_at |
AT2G28570
|
unknown |
0.268 |
0.257 |
| 253 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.268 |
-0.178 |
| 254 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.268 |
-0.733 |
| 255 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.268 |
0.055 |
| 256 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
0.268 |
0.111 |
| 257 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.267 |
0.585 |
| 258 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.267 |
0.481 |
| 259 |
262719_at |
AT1G43590
|
unknown |
0.266 |
-0.296 |
| 260 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.265 |
0.319 |
| 261 |
254077_at |
AT4G25640
|
ATDTX35, detoxifying efflux carrier 35, DTX35, detoxifying efflux carrier 35, FFT, FLOWER FLAVONOID TRANSPORTER |
0.264 |
0.417 |
| 262 |
263032_at |
AT1G23850
|
unknown |
0.263 |
-0.035 |
| 263 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.263 |
0.286 |
| 264 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.261 |
-0.463 |
| 265 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.261 |
-0.445 |
| 266 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.261 |
-0.167 |
| 267 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.261 |
-0.593 |
| 268 |
267178_at |
AT2G37750
|
unknown |
0.261 |
-0.416 |
| 269 |
258581_at |
AT3G04160
|
unknown |
0.259 |
-0.178 |
| 270 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.259 |
0.035 |
| 271 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.258 |
-0.364 |
| 272 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
0.258 |
-0.930 |
| 273 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.258 |
0.149 |
| 274 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.258 |
-0.935 |
| 275 |
258921_at |
AT3G10500
|
ANAC053, NAC domain containing protein 53, NAC053, NAC domain containing protein 53, NTL4, NAC with transmembrane motif 1-like 4 |
0.257 |
-0.077 |
| 276 |
248248_at |
AT5G53120
|
ATSPDS3, SPERMIDINE SYNTHASE 3, SPDS3, spermidine synthase 3, SPMS |
0.255 |
0.742 |
| 277 |
256453_at |
AT1G75270
|
DHAR2, dehydroascorbate reductase 2 |
0.255 |
0.092 |
| 278 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.254 |
-0.420 |
| 279 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
0.254 |
0.287 |
| 280 |
253399_at |
AT4G32850
|
nPAP, nuclear poly(a) polymerase, PAP(IV), poly(A) polymerase IV, PAPS4, poly(A) polymerase 4 |
0.253 |
-0.172 |
| 281 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
0.252 |
-0.662 |
| 282 |
264909_at |
AT2G17300
|
unknown |
0.252 |
-0.740 |
| 283 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.252 |
0.634 |
| 284 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.251 |
0.118 |
| 285 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.251 |
-0.570 |
| 286 |
264580_at |
AT1G05340
|
unknown |
0.251 |
-1.243 |
| 287 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.251 |
-0.513 |
| 288 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.250 |
-0.214 |
| 289 |
260875_at |
AT1G21410
|
SKP2A |
0.250 |
0.330 |
| 290 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.250 |
-0.852 |
| 291 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.250 |
-0.470 |
| 292 |
247464_at |
AT5G62070
|
IQD23, IQ-domain 23 |
0.249 |
0.147 |
| 293 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.249 |
-0.761 |
| 294 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
0.249 |
0.790 |
| 295 |
253962_at |
AT4G26460
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.248 |
-1.156 |
| 296 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.248 |
0.212 |
| 297 |
264255_at |
AT1G09140
|
At-SR30, Serine/Arginine-Rich Protein Splicing Factor 30, ATSRP30, SERINE-ARGININE PROTEIN 30, ATSRP30.1, ATSRP30.2, SR30, Serine/Arginine-Rich Protein Splicing Factor 30 |
0.248 |
1.210 |
| 298 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
0.245 |
0.097 |
| 299 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.245 |
-0.093 |
| 300 |
262391_at |
AT1G49530
|
GGPS6, geranylgeranyl pyrophosphate synthase 6 |
0.245 |
0.512 |
| 301 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.929 |
0.940 |
| 302 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.751 |
-0.003 |
| 303 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.738 |
0.049 |
| 304 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.719 |
-1.133 |
| 305 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.704 |
-0.886 |
| 306 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-0.691 |
0.892 |
| 307 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.632 |
0.059 |
| 308 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.628 |
0.412 |
| 309 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.623 |
-0.600 |
| 310 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.616 |
0.737 |
| 311 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
-0.614 |
0.155 |
| 312 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.612 |
0.031 |
| 313 |
250936_at |
AT5G03120
|
unknown |
-0.607 |
-0.668 |
| 314 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.604 |
-0.128 |
| 315 |
253643_at |
AT4G29780
|
unknown |
-0.586 |
0.234 |
| 316 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
-0.580 |
-0.520 |
| 317 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.561 |
-0.055 |
| 318 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.556 |
-0.385 |
| 319 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.545 |
0.168 |
| 320 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.539 |
0.044 |
| 321 |
267580_at |
AT2G41990
|
unknown |
-0.539 |
-0.539 |
| 322 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.524 |
1.057 |
| 323 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.524 |
1.033 |
| 324 |
246200_at |
AT4G37240
|
unknown |
-0.523 |
1.011 |
| 325 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.514 |
0.850 |
| 326 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.511 |
0.346 |
| 327 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.508 |
1.130 |
| 328 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.506 |
-0.494 |
| 329 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.494 |
-0.661 |
| 330 |
245755_at |
AT1G35210
|
unknown |
-0.490 |
-0.910 |
| 331 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.486 |
0.870 |
| 332 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.481 |
0.042 |
| 333 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.480 |
-0.636 |
| 334 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.470 |
0.938 |
| 335 |
266882_at |
AT2G44670
|
unknown |
-0.469 |
0.554 |
| 336 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.468 |
-0.063 |
| 337 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.467 |
-0.480 |
| 338 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.462 |
-0.242 |
| 339 |
250732_at |
AT5G06480
|
[Immunoglobulin E-set superfamily protein] |
-0.457 |
0.779 |
| 340 |
247214_at |
AT5G64850
|
unknown |
-0.455 |
-0.108 |
| 341 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.454 |
-0.317 |
| 342 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.453 |
1.010 |
| 343 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.452 |
0.587 |
| 344 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
-0.449 |
-0.137 |
| 345 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.447 |
-0.442 |
| 346 |
267623_at |
AT2G39650
|
unknown |
-0.447 |
0.140 |
| 347 |
261658_at |
AT1G50040
|
unknown |
-0.446 |
-0.041 |
| 348 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.445 |
-0.234 |
| 349 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
-0.443 |
1.144 |
| 350 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.442 |
-0.030 |
| 351 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.441 |
0.429 |
| 352 |
253322_at |
AT4G33980
|
unknown |
-0.438 |
-0.998 |
| 353 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.435 |
-0.745 |
| 354 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.432 |
-0.584 |
| 355 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.431 |
-1.134 |
| 356 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.430 |
0.225 |
| 357 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.428 |
-0.599 |
| 358 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.424 |
-0.350 |
| 359 |
250875_at |
AT5G04020
|
[calmodulin binding] |
-0.422 |
-0.726 |
| 360 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.421 |
-1.574 |
| 361 |
248186_at |
AT5G53880
|
unknown |
-0.419 |
0.558 |
| 362 |
256789_at |
AT3G13672
|
SINA2 |
-0.417 |
0.283 |
| 363 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.417 |
-0.236 |
| 364 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.417 |
-0.307 |
| 365 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.414 |
-0.993 |
| 366 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.412 |
0.211 |
| 367 |
267036_at |
AT2G38465
|
unknown |
-0.410 |
-0.896 |
| 368 |
266956_at |
AT2G34510
|
unknown |
-0.407 |
-0.079 |
| 369 |
251142_at |
AT5G01015
|
unknown |
-0.407 |
1.017 |
| 370 |
262797_at |
AT1G20840
|
AtTMT1, TMT1, tonoplast monosaccharide transporter1 |
-0.404 |
-0.202 |
| 371 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.404 |
1.541 |
| 372 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.403 |
0.129 |
| 373 |
267532_at |
AT2G42040
|
unknown |
-0.401 |
0.287 |
| 374 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.401 |
0.181 |
| 375 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
-0.399 |
-0.052 |
| 376 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.395 |
0.370 |
| 377 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.395 |
-1.382 |
| 378 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.393 |
-0.727 |
| 379 |
254043_at |
AT4G25990
|
CIL |
-0.393 |
-0.369 |
| 380 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.391 |
1.405 |
| 381 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.389 |
-0.077 |
| 382 |
259020_at |
AT3G07470
|
unknown |
-0.389 |
0.462 |
| 383 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.388 |
-0.378 |
| 384 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.384 |
-0.037 |
| 385 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.383 |
0.139 |
| 386 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
-0.381 |
1.317 |
| 387 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.381 |
0.242 |
| 388 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.380 |
-0.565 |
| 389 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.380 |
-0.570 |
| 390 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.379 |
-0.542 |
| 391 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.378 |
-0.295 |
| 392 |
264857_at |
AT1G24170
|
GATL8, GALACTURONOSYLTRANSFERASE-LIKE 8, LGT9 |
-0.378 |
0.097 |
| 393 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.377 |
-0.263 |
| 394 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.375 |
-0.082 |
| 395 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.368 |
-0.745 |
| 396 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.368 |
-0.576 |
| 397 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.368 |
1.091 |
| 398 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.367 |
0.429 |
| 399 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.367 |
-0.239 |
| 400 |
256096_at |
AT1G13650
|
unknown |
-0.364 |
-0.683 |
| 401 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.363 |
0.923 |
| 402 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.362 |
0.247 |
| 403 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.359 |
-0.444 |
| 404 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
-0.357 |
-0.331 |
| 405 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.355 |
-0.497 |
| 406 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.352 |
0.773 |
| 407 |
253165_at |
AT4G35320
|
unknown |
-0.351 |
-0.281 |
| 408 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.349 |
0.915 |
| 409 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.348 |
-0.160 |
| 410 |
260655_at |
AT1G19320
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.348 |
-1.210 |
| 411 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.348 |
0.240 |
| 412 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
-0.347 |
-0.514 |
| 413 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.347 |
-0.240 |
| 414 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.346 |
-0.909 |
| 415 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.346 |
0.720 |
| 416 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
-0.345 |
-0.509 |
| 417 |
258468_at |
AT3G06070
|
unknown |
-0.345 |
-1.099 |
| 418 |
247047_at |
AT5G66650
|
unknown |
-0.345 |
-0.786 |
| 419 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.343 |
-0.298 |
| 420 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.343 |
-0.398 |
| 421 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.342 |
-0.776 |
| 422 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.340 |
0.146 |
| 423 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.340 |
-0.823 |
| 424 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-0.340 |
0.840 |
| 425 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.340 |
0.370 |
| 426 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.339 |
-0.084 |
| 427 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-0.339 |
-0.354 |
| 428 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.338 |
-0.189 |
| 429 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
-0.338 |
0.047 |
| 430 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.335 |
0.941 |
| 431 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.334 |
-1.097 |
| 432 |
245254_at |
AT4G14680
|
APS3 |
-0.333 |
0.046 |
| 433 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
-0.333 |
0.936 |
| 434 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.333 |
0.994 |
| 435 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.332 |
0.698 |
| 436 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.332 |
0.909 |
| 437 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.330 |
1.536 |
| 438 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.329 |
-0.819 |
| 439 |
258095_at |
AT3G23610
|
DSPTP1, dual specificity protein phosphatase 1 |
-0.329 |
-0.286 |
| 440 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.328 |
0.279 |
| 441 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.328 |
-0.409 |
| 442 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
-0.328 |
0.008 |
| 443 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.328 |
-0.325 |
| 444 |
264521_at |
AT1G10020
|
unknown |
-0.327 |
0.255 |
| 445 |
253827_at |
AT4G28085
|
unknown |
-0.327 |
-0.942 |
| 446 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.326 |
-0.663 |
| 447 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.326 |
-0.214 |
| 448 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.326 |
1.244 |
| 449 |
264238_at |
AT1G54740
|
unknown |
-0.325 |
-1.118 |
| 450 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.325 |
0.543 |
| 451 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.324 |
-0.746 |
| 452 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.324 |
-0.178 |
| 453 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.324 |
0.081 |
| 454 |
258123_at |
AT3G18040
|
MPK9, MAP kinase 9 |
-0.323 |
-0.174 |
| 455 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.322 |
-0.306 |
| 456 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.321 |
0.564 |
| 457 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.321 |
-0.424 |
| 458 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.321 |
0.667 |
| 459 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.321 |
-0.174 |
| 460 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.320 |
0.130 |
| 461 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
-0.320 |
0.234 |
| 462 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.320 |
0.169 |
| 463 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.320 |
0.392 |
| 464 |
266861_at |
AT2G26830
|
emb1187, embryo defective 1187 |
-0.320 |
-0.026 |
| 465 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.319 |
-0.036 |
| 466 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.319 |
-0.558 |
| 467 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.316 |
1.294 |
| 468 |
249069_at |
AT5G44010
|
unknown |
-0.315 |
-0.246 |
| 469 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.315 |
0.239 |
| 470 |
248865_at |
AT5G46790
|
PYL1, PYR1-like 1, RCAR12, regulatory components of ABA receptor 12 |
-0.315 |
0.629 |
| 471 |
246926_at |
AT5G25240
|
unknown |
-0.314 |
-0.506 |
| 472 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.313 |
-1.379 |
| 473 |
259072_at |
AT3G11700
|
FLA18, FASCICLIN-like arabinogalactan protein 18 precursor |
-0.312 |
0.983 |
| 474 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.311 |
-0.252 |
| 475 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.310 |
-0.684 |
| 476 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.310 |
-0.626 |
| 477 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.309 |
-0.378 |
| 478 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.307 |
0.414 |
| 479 |
246755_at |
AT5G27920
|
[F-box family protein] |
-0.306 |
1.253 |
| 480 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.306 |
0.138 |
| 481 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.306 |
0.136 |
| 482 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.306 |
-0.319 |
| 483 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.305 |
-0.936 |
| 484 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.305 |
-0.925 |
| 485 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.305 |
0.090 |
| 486 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.305 |
-0.202 |
| 487 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.304 |
0.624 |
| 488 |
249123_at |
AT5G43760
|
KCS20, 3-ketoacyl-CoA synthase 20 |
-0.303 |
0.668 |
| 489 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.303 |
-1.116 |
| 490 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.303 |
-0.726 |
| 491 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.303 |
-0.393 |
| 492 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.299 |
0.373 |
| 493 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
-0.299 |
0.148 |
| 494 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.298 |
0.529 |
| 495 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.297 |
0.582 |
| 496 |
267209_at |
AT2G30930
|
unknown |
-0.297 |
-0.540 |
| 497 |
267318_at |
AT2G34770
|
ATFAH1, ARABIDOPSIS FATTY ACID HYDROXYLASE 1, FAH1, fatty acid hydroxylase 1 |
-0.296 |
0.633 |
| 498 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.296 |
-1.148 |
| 499 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.295 |
0.406 |
| 500 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.292 |
-0.320 |
| 501 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.292 |
0.089 |
| 502 |
251636_at |
AT3G57530
|
ATCPK32, CDPK32, CPK32, calcium-dependent protein kinase 32 |
-0.292 |
0.279 |
| 503 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.291 |
-0.140 |
| 504 |
265327_at |
AT2G18210
|
unknown |
-0.291 |
-0.269 |
| 505 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.289 |
-0.567 |
| 506 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.288 |
-0.029 |
| 507 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
-0.288 |
-0.006 |
| 508 |
263606_at |
AT2G16280
|
KCS9, 3-ketoacyl-CoA synthase 9 |
-0.288 |
0.305 |
| 509 |
246528_at |
AT5G15640
|
[Mitochondrial substrate carrier family protein] |
-0.287 |
0.712 |
| 510 |
251646_at |
AT3G57780
|
unknown |
-0.287 |
-0.741 |
| 511 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.286 |
0.417 |
| 512 |
264449_at |
AT1G27460
|
NPGR1, no pollen germination related 1 |
-0.286 |
0.773 |
| 513 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.286 |
0.033 |
| 514 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.286 |
-0.035 |
| 515 |
263795_at |
AT2G24610
|
ATCNGC14, cyclic nucleotide-gated channel 14, CNGC14, cyclic nucleotide-gated channel 14 |
-0.284 |
0.043 |
| 516 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.284 |
-0.102 |
| 517 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
-0.284 |
0.141 |
| 518 |
254363_at |
AT4G22010
|
sks4, SKU5 similar 4 |
-0.282 |
0.483 |
| 519 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.281 |
-0.379 |
| 520 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.280 |
-0.024 |
| 521 |
254487_at |
AT4G20780
|
CML42, calmodulin like 42 |
-0.278 |
-0.318 |
| 522 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.278 |
0.503 |
| 523 |
245209_at |
AT5G12340
|
unknown |
-0.276 |
-1.125 |
| 524 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.276 |
0.292 |
| 525 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.276 |
-0.114 |
| 526 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.275 |
-0.777 |
| 527 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
-0.272 |
0.394 |
| 528 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.272 |
0.516 |
| 529 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.272 |
-1.111 |
| 530 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.272 |
-0.334 |
| 531 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.272 |
0.439 |
| 532 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.271 |
-0.157 |
| 533 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.271 |
0.248 |
| 534 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.271 |
0.067 |
| 535 |
252273_at |
AT3G49660
|
AtWDR5a, WDR5a, human WDR5 (WD40 repeat) homolog a |
-0.270 |
1.036 |
| 536 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.270 |
0.601 |
| 537 |
261292_at |
AT1G36940
|
unknown |
-0.269 |
-0.411 |
| 538 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.269 |
0.958 |
| 539 |
259523_at |
AT1G12500
|
[Nucleotide-sugar transporter family protein] |
-0.268 |
0.882 |
| 540 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.268 |
0.152 |
| 541 |
259856_at |
AT1G68440
|
unknown |
-0.268 |
0.381 |
| 542 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
-0.267 |
-0.315 |
| 543 |
251468_at |
AT3G59290
|
[ENTH/VHS family protein] |
-0.266 |
-0.404 |
| 544 |
251413_at |
AT3G60320
|
unknown |
-0.266 |
1.328 |
| 545 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.265 |
-0.212 |
| 546 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.265 |
0.054 |
| 547 |
249985_at |
AT5G18500
|
[Protein kinase superfamily protein] |
-0.264 |
1.105 |
| 548 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.264 |
-0.029 |
| 549 |
257670_at |
AT3G20340
|
unknown |
-0.264 |
-0.420 |
| 550 |
250009_at |
AT5G18440
|
AtNUFIP, NUFIP, nuclear FMRP-interacting protein |
-0.264 |
0.129 |
| 551 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.264 |
-0.748 |
| 552 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
-0.263 |
0.153 |
| 553 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.262 |
0.318 |
| 554 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.261 |
0.561 |
| 555 |
264576_at |
AT1G05360
|
KMS2, Killing Me Slowly 2 |
-0.261 |
0.200 |
| 556 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.260 |
0.280 |
| 557 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
0.204 |
| 558 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
-0.260 |
0.011 |
| 559 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.259 |
-0.848 |
| 560 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.259 |
-0.834 |
| 561 |
263121_at |
AT1G78530
|
[Protein kinase superfamily protein] |
-0.259 |
-0.281 |
| 562 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.258 |
0.579 |
| 563 |
254815_at |
AT4G12420
|
SKU5 |
-0.257 |
0.506 |
| 564 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.256 |
-0.292 |
| 565 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.256 |
-0.166 |
| 566 |
247394_at |
AT5G62865
|
unknown |
-0.256 |
-0.521 |
| 567 |
255733_at |
AT1G25400
|
unknown |
-0.255 |
-0.713 |
| 568 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
-0.255 |
-0.120 |
| 569 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.255 |
0.925 |
| 570 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.255 |
0.455 |
| 571 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.255 |
0.466 |
| 572 |
257202_at |
AT3G23750
|
BARK1, BAK1-Associating Receptor-Like Kinase 1 |
-0.255 |
0.816 |
| 573 |
261818_at |
AT1G11390
|
[Protein kinase superfamily protein] |
-0.254 |
0.308 |
| 574 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
-0.254 |
0.695 |
| 575 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.254 |
0.592 |
| 576 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.253 |
0.245 |
| 577 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.253 |
-0.139 |
| 578 |
250980_at |
AT5G03130
|
unknown |
-0.252 |
0.334 |
| 579 |
246217_at |
AT4G36920
|
AP2, APETALA 2, AtAP2, FL1, FLOWER 1, FLO2, FLORAL MUTANT 2 |
-0.252 |
0.489 |
| 580 |
263249_at |
AT2G31360
|
ADS2, 16:0delta9 desaturase 2, AtADS2 |
-0.252 |
0.910 |
| 581 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.251 |
-0.897 |
| 582 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.251 |
0.113 |
| 583 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.251 |
-0.684 |
| 584 |
253891_at |
AT4G27720
|
[Major facilitator superfamily protein] |
-0.250 |
0.708 |
| 585 |
247486_at |
AT5G62140
|
unknown |
-0.250 |
-0.354 |
| 586 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.250 |
-0.725 |
| 587 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
-0.249 |
0.695 |
| 588 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.249 |
-0.851 |
| 589 |
255808_at |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
-0.249 |
0.640 |
| 590 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.248 |
-0.164 |
| 591 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
-0.248 |
0.802 |
| 592 |
266732_at |
AT2G03240
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.248 |
0.417 |
| 593 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
-0.248 |
-0.981 |
| 594 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.247 |
-0.768 |
| 595 |
265544_at |
AT2G28260
|
ATCNGC15, cyclic nucleotide-gated channel 15, CNGC15, cyclic nucleotide-gated channel 15 |
-0.247 |
-0.774 |
| 596 |
256021_at |
AT1G58270
|
ZW9 |
-0.246 |
-0.195 |
| 597 |
253454_at |
AT4G31875
|
unknown |
-0.246 |
-0.121 |
| 598 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.245 |
0.343 |
| 599 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
-0.245 |
-0.724 |
| 600 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.245 |
-0.988 |