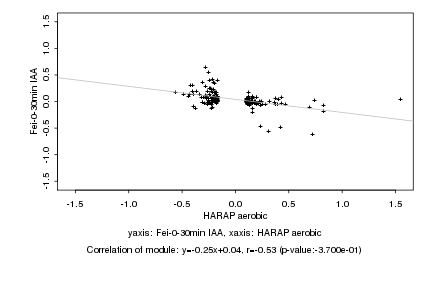
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
HARAP aerobic |
Log2 signal ratio
Fei-0-30min IAA |
| 1 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.543 |
0.050 |
| 2 |
247024_at |
AT5G66985
|
unknown |
0.824 |
-0.062 |
| 3 |
249384_at |
AT5G39890
|
unknown |
0.823 |
-0.171 |
| 4 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.735 |
0.033 |
| 5 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.722 |
-0.617 |
| 6 |
250152_at |
AT5G15120
|
unknown |
0.685 |
-0.102 |
| 7 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.463 |
-0.046 |
| 8 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.425 |
0.080 |
| 9 |
252882_at |
AT4G39675
|
unknown |
0.425 |
-0.026 |
| 10 |
261567_at |
AT1G33055
|
unknown |
0.418 |
-0.473 |
| 11 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.396 |
0.049 |
| 12 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.394 |
-0.047 |
| 13 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.370 |
-0.051 |
| 14 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.366 |
0.057 |
| 15 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.363 |
-0.003 |
| 16 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.311 |
0.006 |
| 17 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.309 |
-0.558 |
| 18 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.274 |
-0.040 |
| 19 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.247 |
-0.040 |
| 20 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.235 |
0.001 |
| 21 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.232 |
-0.063 |
| 22 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.231 |
-0.009 |
| 23 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.226 |
-0.468 |
| 24 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.220 |
0.009 |
| 25 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.212 |
-0.021 |
| 26 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.211 |
-0.035 |
| 27 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.209 |
-0.029 |
| 28 |
253165_at |
AT4G35320
|
unknown |
0.199 |
-0.026 |
| 29 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.195 |
0.085 |
| 30 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
0.193 |
-0.054 |
| 31 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.192 |
0.085 |
| 32 |
246385_at |
AT1G77390
|
CYCA1, CYCLIN A1, CYCA1;2, CYCLIN A1;2, DYP, TAM, TARDY ASYNCHRONOUS MEIOSIS |
0.183 |
0.001 |
| 33 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
0.181 |
-0.007 |
| 34 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.175 |
-0.032 |
| 35 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.173 |
0.015 |
| 36 |
253050_at |
AT4G37450
|
AGP18, arabinogalactan protein 18, ATAGP18 |
0.170 |
-0.022 |
| 37 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
0.156 |
0.080 |
| 38 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.155 |
-0.037 |
| 39 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.155 |
-0.197 |
| 40 |
260686_at |
AT1G17620
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.155 |
0.005 |
| 41 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.154 |
0.112 |
| 42 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.154 |
-0.131 |
| 43 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.153 |
0.061 |
| 44 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.153 |
-0.009 |
| 45 |
254611_at |
AT4G19095
|
unknown |
0.151 |
0.005 |
| 46 |
250201_at |
AT5G14230
|
unknown |
0.150 |
0.031 |
| 47 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.148 |
-0.012 |
| 48 |
265025_at |
AT1G24575
|
unknown |
0.144 |
-0.006 |
| 49 |
262197_at |
AT1G53910
|
RAP2.12, related to AP2 12 |
0.142 |
0.002 |
| 50 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
0.139 |
0.019 |
| 51 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.137 |
-0.003 |
| 52 |
267470_at |
AT2G30490
|
ATC4H, CINNAMATE 4-HYDROXYLASE, C4H, cinnamate-4-hydroxylase, CYP73A5, REF3, REDUCED EPRDERMAL FLUORESCENCE 3 |
0.134 |
-0.014 |
| 53 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.132 |
-0.033 |
| 54 |
251394_at |
AT3G60900
|
FLA10, FASCICLIN-like arabinogalactan-protein 10 |
0.131 |
-0.066 |
| 55 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
0.129 |
-0.004 |
| 56 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.129 |
-0.045 |
| 57 |
265171_at |
AT1G23790
|
unknown |
0.127 |
0.033 |
| 58 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
0.126 |
-0.042 |
| 59 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.126 |
0.059 |
| 60 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
0.125 |
0.015 |
| 61 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
0.122 |
-0.021 |
| 62 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
0.122 |
-0.025 |
| 63 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.121 |
0.031 |
| 64 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.120 |
0.174 |
| 65 |
253931_at |
AT4G26770
|
[Phosphatidate cytidylyltransferase family protein] |
0.119 |
0.025 |
| 66 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.117 |
-0.048 |
| 67 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
0.116 |
-0.015 |
| 68 |
257813_at |
AT3G25100
|
CDC45, cell division cycle 45 |
0.115 |
0.037 |
| 69 |
263971_at |
AT2G42800
|
AtRLP29, receptor like protein 29, RLP29, receptor like protein 29 |
0.114 |
0.065 |
| 70 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.114 |
0.106 |
| 71 |
247301_at |
AT5G63920
|
AtTOP3alpha, TOP3A, topoisomerase 3alpha |
0.111 |
0.038 |
| 72 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
0.110 |
-0.031 |
| 73 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.110 |
0.067 |
| 74 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.110 |
0.020 |
| 75 |
261876_at |
AT1G50590
|
[RmlC-like cupins superfamily protein] |
0.109 |
-0.034 |
| 76 |
246555_at |
AT5G15470
|
GAUT14, galacturonosyltransferase 14 |
0.108 |
-0.034 |
| 77 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
0.107 |
-0.048 |
| 78 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.107 |
0.012 |
| 79 |
257697_at |
AT3G12700
|
NANA, NANA |
0.105 |
-0.014 |
| 80 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.104 |
0.031 |
| 81 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
0.104 |
0.012 |
| 82 |
251223_at |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
0.103 |
0.059 |
| 83 |
249821_at |
AT5G23690
|
[Polynucleotide adenylyltransferase family protein] |
0.103 |
0.004 |
| 84 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.103 |
0.014 |
| 85 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.102 |
0.014 |
| 86 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.101 |
0.020 |
| 87 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
0.101 |
0.036 |
| 88 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
0.101 |
0.033 |
| 89 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.101 |
-0.010 |
| 90 |
257618_at |
AT3G24715
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
0.100 |
0.045 |
| 91 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
-0.571 |
0.186 |
| 92 |
245755_at |
AT1G35210
|
unknown |
-0.488 |
0.134 |
| 93 |
256442_at |
AT3G10930
|
unknown |
-0.442 |
0.106 |
| 94 |
253643_at |
AT4G29780
|
unknown |
-0.439 |
0.145 |
| 95 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.425 |
0.312 |
| 96 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.408 |
0.318 |
| 97 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.406 |
0.199 |
| 98 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.400 |
-0.081 |
| 99 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.394 |
0.143 |
| 100 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.376 |
-0.114 |
| 101 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.370 |
0.203 |
| 102 |
253044_at |
AT4G37290
|
unknown |
-0.344 |
0.139 |
| 103 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.320 |
0.088 |
| 104 |
246923_at |
AT5G25100
|
[Endomembrane protein 70 protein family] |
-0.317 |
-0.001 |
| 105 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.317 |
0.363 |
| 106 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.312 |
-0.017 |
| 107 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.305 |
0.092 |
| 108 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.290 |
-0.029 |
| 109 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.287 |
0.290 |
| 110 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.285 |
0.644 |
| 111 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.284 |
0.122 |
| 112 |
253322_at |
AT4G33980
|
unknown |
-0.276 |
0.094 |
| 113 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.276 |
0.072 |
| 114 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.274 |
0.110 |
| 115 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.268 |
-0.041 |
| 116 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.267 |
0.200 |
| 117 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
-0.265 |
-0.012 |
| 118 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.261 |
0.024 |
| 119 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.255 |
0.556 |
| 120 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.254 |
0.059 |
| 121 |
263182_at |
AT1G05575
|
unknown |
-0.252 |
0.407 |
| 122 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.252 |
0.254 |
| 123 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.248 |
-0.022 |
| 124 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.246 |
-0.005 |
| 125 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.244 |
0.136 |
| 126 |
250098_at |
AT5G17350
|
unknown |
-0.242 |
0.104 |
| 127 |
267623_at |
AT2G39650
|
unknown |
-0.241 |
0.253 |
| 128 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.231 |
0.021 |
| 129 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.230 |
-0.114 |
| 130 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.229 |
0.025 |
| 131 |
250796_at |
AT5G05300
|
unknown |
-0.228 |
-0.038 |
| 132 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.226 |
0.058 |
| 133 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-0.226 |
0.190 |
| 134 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.225 |
0.241 |
| 135 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.224 |
0.013 |
| 136 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.223 |
0.085 |
| 137 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
-0.222 |
-0.008 |
| 138 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.222 |
-0.008 |
| 139 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.222 |
0.425 |
| 140 |
265184_at |
AT1G23710
|
unknown |
-0.220 |
-0.007 |
| 141 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
-0.220 |
-0.108 |
| 142 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
-0.218 |
0.081 |
| 143 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.217 |
0.187 |
| 144 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.213 |
0.364 |
| 145 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.210 |
0.042 |
| 146 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.210 |
0.033 |
| 147 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.206 |
0.232 |
| 148 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-0.206 |
0.079 |
| 149 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.206 |
0.095 |
| 150 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.206 |
0.017 |
| 151 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
-0.204 |
0.026 |
| 152 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.203 |
0.354 |
| 153 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.202 |
0.080 |
| 154 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.200 |
0.181 |
| 155 |
262452_at |
AT1G11210
|
unknown |
-0.199 |
0.003 |
| 156 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.198 |
0.098 |
| 157 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
-0.197 |
0.085 |
| 158 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.192 |
0.176 |
| 159 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.189 |
0.059 |
| 160 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.189 |
0.083 |
| 161 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.185 |
-0.000 |
| 162 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
-0.185 |
0.013 |
| 163 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.183 |
0.178 |
| 164 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
-0.182 |
0.024 |
| 165 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.180 |
0.141 |
| 166 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.179 |
-0.020 |
| 167 |
249896_at |
AT5G22530
|
unknown |
-0.178 |
0.029 |
| 168 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.178 |
0.042 |
| 169 |
253859_at |
AT4G27657
|
unknown |
-0.177 |
0.061 |
| 170 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.177 |
0.403 |
| 171 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.176 |
0.021 |
| 172 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.174 |
0.052 |
| 173 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
-0.174 |
0.092 |
| 174 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.172 |
0.018 |
| 175 |
259479_at |
AT1G19020
|
unknown |
-0.172 |
0.105 |
| 176 |
246584_at |
AT5G14730
|
unknown |
-0.170 |
0.035 |
| 177 |
252474_at |
AT3G46620
|
unknown |
-0.170 |
0.003 |
| 178 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.170 |
0.109 |
| 179 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.169 |
0.030 |