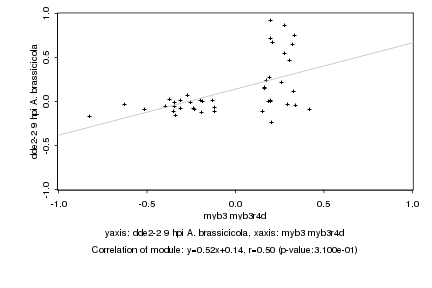
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
myb3 myb3r4d |
Log2 signal ratio
dde2-2 9 hpi A. brassicicola |
| 1 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.417 |
-0.081 |
| 2 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.337 |
-0.039 |
| 3 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.329 |
0.752 |
| 4 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.324 |
0.125 |
| 5 |
249896_at |
AT5G22530
|
unknown |
0.322 |
0.652 |
| 6 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.303 |
0.471 |
| 7 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.292 |
-0.026 |
| 8 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.275 |
0.551 |
| 9 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.274 |
0.869 |
| 10 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.257 |
0.225 |
| 11 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.205 |
0.672 |
| 12 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.202 |
-0.227 |
| 13 |
259697_at |
AT1G68920
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.197 |
0.002 |
| 14 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.196 |
0.931 |
| 15 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.196 |
0.022 |
| 16 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.193 |
0.726 |
| 17 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.192 |
0.282 |
| 18 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.185 |
0.009 |
| 19 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.173 |
0.240 |
| 20 |
251277_at |
AT3G61760
|
ADL1B, DYNAMIN-like 1B, DL1B, DYNAMIN-like 1B |
0.163 |
0.155 |
| 21 |
265189_at |
AT1G23840
|
unknown |
0.159 |
0.166 |
| 22 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.152 |
-0.112 |
| 23 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.828 |
-0.170 |
| 24 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.631 |
-0.029 |
| 25 |
252089_at |
AT3G52110
|
unknown |
-0.519 |
-0.087 |
| 26 |
262558_at |
AT1G31335
|
unknown |
-0.396 |
-0.049 |
| 27 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.378 |
0.027 |
| 28 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.351 |
-0.109 |
| 29 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.349 |
-0.001 |
| 30 |
252077_at |
AT3G51720
|
unknown |
-0.348 |
-0.053 |
| 31 |
258480_at |
AT3G02640
|
unknown |
-0.339 |
-0.157 |
| 32 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.316 |
-0.078 |
| 33 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.313 |
0.022 |
| 34 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
-0.276 |
0.074 |
| 35 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.258 |
-0.001 |
| 36 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.238 |
-0.077 |
| 37 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.235 |
-0.089 |
| 38 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.200 |
0.017 |
| 39 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.194 |
-0.116 |
| 40 |
250979_at |
AT5G03090
|
unknown |
-0.189 |
0.009 |
| 41 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.131 |
0.014 |
| 42 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.124 |
-0.060 |
| 43 |
248265_at |
AT5G53430
|
ATX5, SDG29, SET domain group 29, SET29 |
-0.122 |
-0.104 |