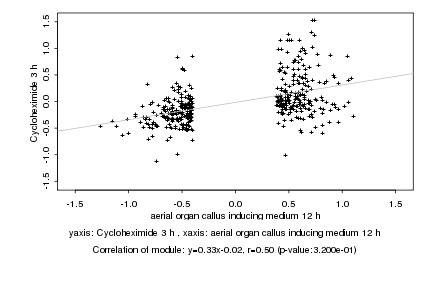
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
aerial organ callus inducing medium 12 h |
Log2 signal ratio
Cycloheximide 3 h |
| 1 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.099 |
-0.263 |
| 2 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.079 |
0.446 |
| 3 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.051 |
0.412 |
| 4 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.050 |
-0.009 |
| 5 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
1.040 |
0.854 |
| 6 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.021 |
-0.075 |
| 7 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.962 |
-0.379 |
| 8 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.962 |
-0.132 |
| 9 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.957 |
0.353 |
| 10 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.932 |
-0.098 |
| 11 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.927 |
0.456 |
| 12 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.921 |
-0.042 |
| 13 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.913 |
0.502 |
| 14 |
251012_at |
AT5G02580
|
unknown |
0.905 |
-0.042 |
| 15 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.886 |
0.883 |
| 16 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.883 |
-0.168 |
| 17 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.874 |
-0.380 |
| 18 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.848 |
-0.003 |
| 19 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.844 |
0.392 |
| 20 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.829 |
0.343 |
| 21 |
259735_at |
AT1G64405
|
unknown |
0.818 |
-0.135 |
| 22 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.813 |
-0.589 |
| 23 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.811 |
0.027 |
| 24 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.811 |
-0.209 |
| 25 |
260668_at |
AT1G19530
|
unknown |
0.807 |
-0.438 |
| 26 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.800 |
0.078 |
| 27 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.793 |
-0.119 |
| 28 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.787 |
0.364 |
| 29 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.772 |
0.678 |
| 30 |
245076_at |
AT2G23170
|
GH3.3 |
0.764 |
0.887 |
| 31 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.750 |
0.030 |
| 32 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.748 |
-0.482 |
| 33 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.745 |
0.056 |
| 34 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.737 |
1.244 |
| 35 |
258397_at |
AT3G15357
|
unknown |
0.731 |
-0.172 |
| 36 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.731 |
1.524 |
| 37 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.719 |
1.542 |
| 38 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.717 |
0.409 |
| 39 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.716 |
1.022 |
| 40 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.714 |
-0.295 |
| 41 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.712 |
-0.231 |
| 42 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.711 |
-0.065 |
| 43 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.706 |
1.306 |
| 44 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.705 |
0.243 |
| 45 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.704 |
-0.566 |
| 46 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
0.698 |
0.015 |
| 47 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.693 |
0.653 |
| 48 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.692 |
-0.263 |
| 49 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.690 |
-0.140 |
| 50 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.687 |
-0.032 |
| 51 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.684 |
0.021 |
| 52 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.684 |
-0.132 |
| 53 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.684 |
0.298 |
| 54 |
250709_at |
AT5G06080
|
LBD33, LOB domain-containing protein 33 |
0.681 |
-0.024 |
| 55 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.668 |
0.382 |
| 56 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.660 |
0.191 |
| 57 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.655 |
0.313 |
| 58 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.654 |
0.449 |
| 59 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.654 |
0.906 |
| 60 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.652 |
0.526 |
| 61 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.650 |
0.607 |
| 62 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.650 |
-0.126 |
| 63 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.646 |
-0.049 |
| 64 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.643 |
0.964 |
| 65 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.643 |
0.051 |
| 66 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.641 |
0.131 |
| 67 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.638 |
0.089 |
| 68 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.636 |
0.692 |
| 69 |
260553_at |
AT2G41800
|
unknown |
0.633 |
0.345 |
| 70 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.632 |
-0.083 |
| 71 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
0.631 |
-0.326 |
| 72 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.627 |
1.010 |
| 73 |
262805_at |
AT1G20900
|
AHL27, AT-hook motif nuclear-localized protein 27, ESC, ESCAROLA, ORE7, ORESARA 7 |
0.627 |
-0.069 |
| 74 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.625 |
0.139 |
| 75 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.623 |
0.443 |
| 76 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.621 |
0.454 |
| 77 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.621 |
0.242 |
| 78 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.615 |
0.803 |
| 79 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.615 |
-0.182 |
| 80 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.612 |
-0.566 |
| 81 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.609 |
0.142 |
| 82 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.608 |
-0.122 |
| 83 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.605 |
-0.531 |
| 84 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.604 |
-0.302 |
| 85 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.604 |
-0.017 |
| 86 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.600 |
0.959 |
| 87 |
246929_at |
AT5G25210
|
unknown |
0.600 |
0.129 |
| 88 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.597 |
-0.047 |
| 89 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.594 |
-0.011 |
| 90 |
262452_at |
AT1G11210
|
unknown |
0.594 |
0.342 |
| 91 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.593 |
0.020 |
| 92 |
266017_at |
AT2G18690
|
unknown |
0.591 |
0.476 |
| 93 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.591 |
1.155 |
| 94 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.589 |
0.171 |
| 95 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.588 |
0.857 |
| 96 |
257643_at |
AT3G25730
|
EDF3, ethylene response DNA binding factor 3 |
0.586 |
0.603 |
| 97 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.585 |
0.054 |
| 98 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.584 |
0.353 |
| 99 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.584 |
0.748 |
| 100 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.582 |
-0.312 |
| 101 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.578 |
-0.080 |
| 102 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.577 |
0.096 |
| 103 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.576 |
0.412 |
| 104 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.570 |
-0.170 |
| 105 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.570 |
0.074 |
| 106 |
256114_at |
AT1G16850
|
unknown |
0.568 |
-0.079 |
| 107 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.568 |
-0.286 |
| 108 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.561 |
0.773 |
| 109 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.558 |
0.158 |
| 110 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.557 |
0.053 |
| 111 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.556 |
0.097 |
| 112 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.550 |
0.521 |
| 113 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.545 |
0.774 |
| 114 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.544 |
-0.012 |
| 115 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.543 |
-0.135 |
| 116 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.542 |
0.471 |
| 117 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.541 |
0.322 |
| 118 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.539 |
0.746 |
| 119 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.538 |
-0.068 |
| 120 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.538 |
0.616 |
| 121 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.536 |
0.401 |
| 122 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.533 |
-0.078 |
| 123 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.521 |
-0.347 |
| 124 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.521 |
0.181 |
| 125 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.519 |
1.162 |
| 126 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
0.518 |
-0.113 |
| 127 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.511 |
0.121 |
| 128 |
245064_at |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
0.510 |
-0.099 |
| 129 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.510 |
-0.203 |
| 130 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
0.510 |
-0.141 |
| 131 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
0.505 |
-0.027 |
| 132 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.504 |
0.298 |
| 133 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.504 |
1.154 |
| 134 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.501 |
0.038 |
| 135 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.493 |
-0.242 |
| 136 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.491 |
0.080 |
| 137 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.490 |
0.008 |
| 138 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.489 |
0.026 |
| 139 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.488 |
1.273 |
| 140 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
0.485 |
0.084 |
| 141 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.484 |
1.158 |
| 142 |
253047_at |
AT4G37295
|
unknown |
0.484 |
0.137 |
| 143 |
263894_at |
AT2G21910
|
CYP96A5, cytochrome P450, family 96, subfamily A, polypeptide 5 |
0.483 |
0.923 |
| 144 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.482 |
0.088 |
| 145 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.481 |
0.031 |
| 146 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.479 |
0.647 |
| 147 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.472 |
0.017 |
| 148 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.472 |
-0.074 |
| 149 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.472 |
-0.145 |
| 150 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.467 |
0.213 |
| 151 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.467 |
0.201 |
| 152 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.466 |
-0.032 |
| 153 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.465 |
-0.145 |
| 154 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.464 |
0.528 |
| 155 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.464 |
0.332 |
| 156 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.464 |
-1.006 |
| 157 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.464 |
0.081 |
| 158 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.464 |
0.037 |
| 159 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
0.462 |
0.216 |
| 160 |
249941_at |
AT5G22270
|
unknown |
0.462 |
0.173 |
| 161 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.461 |
0.020 |
| 162 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.457 |
-0.101 |
| 163 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.454 |
-0.343 |
| 164 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
0.451 |
0.561 |
| 165 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.451 |
0.200 |
| 166 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.451 |
-0.111 |
| 167 |
247878_at |
AT5G57760
|
unknown |
0.450 |
-0.458 |
| 168 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
0.446 |
-0.056 |
| 169 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.442 |
-0.212 |
| 170 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.440 |
-0.142 |
| 171 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.438 |
0.022 |
| 172 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.437 |
-0.216 |
| 173 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.435 |
0.657 |
| 174 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.434 |
-0.110 |
| 175 |
258878_at |
AT3G03170
|
unknown |
0.434 |
-0.080 |
| 176 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.433 |
-0.242 |
| 177 |
265479_at |
AT2G15760
|
unknown |
0.432 |
0.420 |
| 178 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.430 |
-0.077 |
| 179 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.430 |
0.059 |
| 180 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.429 |
0.005 |
| 181 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
0.428 |
-0.047 |
| 182 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.428 |
0.249 |
| 183 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.426 |
-0.025 |
| 184 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.426 |
0.995 |
| 185 |
252822_at |
AT4G39955
|
[alpha/beta-Hydrolases superfamily protein] |
0.426 |
-0.050 |
| 186 |
252118_at |
AT3G51400
|
unknown |
0.424 |
0.046 |
| 187 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.423 |
-0.052 |
| 188 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.422 |
-0.193 |
| 189 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.422 |
0.213 |
| 190 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
0.419 |
0.602 |
| 191 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.419 |
0.119 |
| 192 |
245352_at |
AT4G15490
|
UGT84A3 |
0.418 |
-0.063 |
| 193 |
246584_at |
AT5G14730
|
unknown |
0.417 |
1.162 |
| 194 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.417 |
0.730 |
| 195 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.415 |
0.106 |
| 196 |
267036_at |
AT2G38465
|
unknown |
0.412 |
0.603 |
| 197 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.410 |
0.573 |
| 198 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.407 |
-0.093 |
| 199 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.404 |
0.007 |
| 200 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.404 |
0.016 |
| 201 |
245439_at |
AT4G16670
|
unknown |
0.401 |
0.009 |
| 202 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.400 |
-0.016 |
| 203 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.399 |
0.023 |
| 204 |
262607_at |
AT1G13990
|
unknown |
0.399 |
0.099 |
| 205 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.398 |
-0.197 |
| 206 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.397 |
-0.409 |
| 207 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.396 |
0.980 |
| 208 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.396 |
-0.004 |
| 209 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.396 |
0.033 |
| 210 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.393 |
0.245 |
| 211 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.392 |
0.007 |
| 212 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.391 |
0.078 |
| 213 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.390 |
-0.070 |
| 214 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.383 |
0.113 |
| 215 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.382 |
-0.059 |
| 216 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-1.271 |
-0.454 |
| 217 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-1.156 |
-0.372 |
| 218 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-1.119 |
-0.458 |
| 219 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-1.061 |
-0.624 |
| 220 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.017 |
-0.331 |
| 221 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-1.009 |
-0.597 |
| 222 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.940 |
-0.278 |
| 223 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.938 |
-0.242 |
| 224 |
256096_at |
AT1G13650
|
unknown |
-0.891 |
-0.385 |
| 225 |
265892_at |
AT2G15020
|
unknown |
-0.876 |
-0.091 |
| 226 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.866 |
-0.284 |
| 227 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.861 |
-0.472 |
| 228 |
254145_at |
AT4G24700
|
unknown |
-0.851 |
-0.329 |
| 229 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.835 |
-0.282 |
| 230 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.834 |
-0.401 |
| 231 |
264636_at |
AT1G65490
|
unknown |
-0.826 |
0.324 |
| 232 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.819 |
-0.708 |
| 233 |
256603_at |
AT3G28270
|
unknown |
-0.819 |
-0.428 |
| 234 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.808 |
-0.480 |
| 235 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.805 |
-0.325 |
| 236 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.803 |
-0.053 |
| 237 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.781 |
-0.643 |
| 238 |
263674_at |
AT2G04790
|
unknown |
-0.779 |
-0.504 |
| 239 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.778 |
0.000 |
| 240 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.778 |
-0.424 |
| 241 |
249932_at |
AT5G22390
|
unknown |
-0.773 |
-0.201 |
| 242 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.770 |
-0.379 |
| 243 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.756 |
-0.431 |
| 244 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.748 |
-1.125 |
| 245 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.748 |
-0.461 |
| 246 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.747 |
-0.440 |
| 247 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.741 |
-0.260 |
| 248 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.730 |
-0.460 |
| 249 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.727 |
-0.073 |
| 250 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.699 |
-0.215 |
| 251 |
263410_at |
AT2G04039
|
unknown |
-0.684 |
-0.280 |
| 252 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.678 |
-0.136 |
| 253 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.677 |
-0.263 |
| 254 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.677 |
-0.102 |
| 255 |
247214_at |
AT5G64850
|
unknown |
-0.676 |
-0.147 |
| 256 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.669 |
-0.275 |
| 257 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.668 |
-0.147 |
| 258 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.665 |
0.036 |
| 259 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.661 |
-0.311 |
| 260 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.660 |
-0.088 |
| 261 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.656 |
-0.179 |
| 262 |
246142_at |
AT5G19970
|
unknown |
-0.649 |
-0.481 |
| 263 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.648 |
0.058 |
| 264 |
264201_at |
AT1G22630
|
unknown |
-0.648 |
-0.284 |
| 265 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.644 |
-0.298 |
| 266 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.643 |
-0.728 |
| 267 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.642 |
-0.140 |
| 268 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.637 |
-0.342 |
| 269 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.635 |
0.063 |
| 270 |
247977_at |
AT5G56850
|
unknown |
-0.632 |
0.004 |
| 271 |
251727_at |
AT3G56290
|
unknown |
-0.631 |
-0.212 |
| 272 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.627 |
-0.315 |
| 273 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.626 |
-0.432 |
| 274 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.622 |
-0.145 |
| 275 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
-0.618 |
-0.018 |
| 276 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.615 |
-0.221 |
| 277 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.615 |
-0.676 |
| 278 |
247486_at |
AT5G62140
|
unknown |
-0.615 |
-0.474 |
| 279 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.606 |
-0.504 |
| 280 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.606 |
-0.226 |
| 281 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.603 |
-0.217 |
| 282 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.603 |
-0.331 |
| 283 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.598 |
-0.313 |
| 284 |
260696_at |
AT1G32520
|
unknown |
-0.597 |
-0.366 |
| 285 |
253305_at |
AT4G33666
|
unknown |
-0.596 |
-0.103 |
| 286 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.595 |
0.268 |
| 287 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.595 |
-0.181 |
| 288 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.594 |
-0.240 |
| 289 |
254642_at |
AT4G18810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.592 |
-0.068 |
| 290 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.592 |
-0.122 |
| 291 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.587 |
-0.218 |
| 292 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.586 |
-0.240 |
| 293 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.585 |
-0.327 |
| 294 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.581 |
-0.239 |
| 295 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.576 |
0.050 |
| 296 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.571 |
0.223 |
| 297 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.570 |
-0.215 |
| 298 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.567 |
-0.454 |
| 299 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.565 |
-0.490 |
| 300 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.560 |
-0.233 |
| 301 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.560 |
-0.160 |
| 302 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.558 |
-0.166 |
| 303 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.558 |
-0.338 |
| 304 |
267063_at |
AT2G41120
|
unknown |
-0.555 |
0.352 |
| 305 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
-0.554 |
-0.157 |
| 306 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.552 |
-0.244 |
| 307 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.549 |
-0.301 |
| 308 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.549 |
0.838 |
| 309 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.546 |
-0.329 |
| 310 |
259460_at |
AT1G44000
|
unknown |
-0.546 |
-0.322 |
| 311 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.544 |
-0.987 |
| 312 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.542 |
0.288 |
| 313 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.541 |
0.241 |
| 314 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.540 |
-0.367 |
| 315 |
264096_at |
AT1G78995
|
unknown |
-0.536 |
-0.195 |
| 316 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.534 |
0.018 |
| 317 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.534 |
-0.192 |
| 318 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.534 |
0.171 |
| 319 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.531 |
-0.202 |
| 320 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.530 |
-0.304 |
| 321 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.528 |
-0.511 |
| 322 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.522 |
-0.088 |
| 323 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.520 |
-0.202 |
| 324 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.518 |
0.273 |
| 325 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.515 |
-0.332 |
| 326 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.514 |
0.143 |
| 327 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.513 |
0.016 |
| 328 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.511 |
-0.422 |
| 329 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.510 |
-0.019 |
| 330 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.509 |
-0.210 |
| 331 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.509 |
-0.027 |
| 332 |
245776_at |
AT1G30260
|
unknown |
-0.508 |
-0.337 |
| 333 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.508 |
0.081 |
| 334 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.506 |
-0.081 |
| 335 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.506 |
-0.290 |
| 336 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.505 |
-0.120 |
| 337 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.505 |
-0.433 |
| 338 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.505 |
0.603 |
| 339 |
253302_at |
AT4G33660
|
unknown |
-0.501 |
-0.519 |
| 340 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.501 |
0.625 |
| 341 |
250696_at |
AT5G06790
|
unknown |
-0.501 |
-0.231 |
| 342 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
-0.497 |
-0.049 |
| 343 |
259275_at |
AT3G01060
|
unknown |
-0.497 |
-0.191 |
| 344 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.494 |
-0.417 |
| 345 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.493 |
-0.478 |
| 346 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.486 |
-0.280 |
| 347 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.483 |
0.594 |
| 348 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.482 |
-0.113 |
| 349 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.481 |
-0.110 |
| 350 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.479 |
-0.068 |
| 351 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.474 |
-0.039 |
| 352 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.473 |
0.191 |
| 353 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.472 |
0.077 |
| 354 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.469 |
-0.119 |
| 355 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.469 |
-0.107 |
| 356 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.468 |
-0.510 |
| 357 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.466 |
-0.425 |
| 358 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.463 |
-0.495 |
| 359 |
262162_at |
AT1G78020
|
unknown |
-0.463 |
-0.448 |
| 360 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.461 |
-0.257 |
| 361 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.460 |
-0.071 |
| 362 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.459 |
-0.530 |
| 363 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.458 |
-0.212 |
| 364 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.458 |
-0.252 |
| 365 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.457 |
-0.284 |
| 366 |
264909_at |
AT2G17300
|
unknown |
-0.456 |
-0.053 |
| 367 |
267344_at |
AT2G44230
|
unknown |
-0.455 |
-0.182 |
| 368 |
258468_at |
AT3G06070
|
unknown |
-0.452 |
-0.541 |
| 369 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.452 |
-0.313 |
| 370 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.449 |
-0.124 |
| 371 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.448 |
-0.307 |
| 372 |
252353_at |
AT3G48200
|
unknown |
-0.448 |
-0.448 |
| 373 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.447 |
-0.283 |
| 374 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.447 |
-0.283 |
| 375 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.446 |
-0.521 |
| 376 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.446 |
-0.256 |
| 377 |
259001_at |
AT3G01960
|
unknown |
-0.444 |
-0.027 |
| 378 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.443 |
-0.290 |
| 379 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.441 |
-0.392 |
| 380 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
-0.441 |
0.114 |
| 381 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.441 |
0.059 |
| 382 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.441 |
-0.129 |
| 383 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.439 |
-0.143 |
| 384 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.439 |
-0.464 |
| 385 |
255304_at |
AT4G04850
|
ATKEA3, KEA3, K+ efflux antiporter 3 |
-0.438 |
-0.176 |
| 386 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.438 |
0.308 |
| 387 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.438 |
-0.373 |
| 388 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.437 |
0.051 |
| 389 |
259856_at |
AT1G68440
|
unknown |
-0.437 |
-0.020 |
| 390 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.436 |
-0.054 |
| 391 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.436 |
-0.286 |
| 392 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.435 |
-0.130 |
| 393 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
-0.435 |
-0.074 |
| 394 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.434 |
-0.126 |
| 395 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
-0.434 |
-0.134 |
| 396 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.434 |
-0.134 |
| 397 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.433 |
-0.222 |
| 398 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.433 |
-0.274 |
| 399 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.432 |
-0.108 |
| 400 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
-0.432 |
-0.022 |
| 401 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
-0.431 |
-0.136 |
| 402 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.431 |
-0.116 |
| 403 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.426 |
-0.211 |
| 404 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.426 |
-0.303 |
| 405 |
250926_at |
AT5G03555
|
AtNCS1, NCS1, nucleobase cation symporter 1 |
-0.424 |
-0.113 |
| 406 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.423 |
-0.366 |
| 407 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.423 |
-0.276 |
| 408 |
250128_at |
AT5G16540
|
ZFN3, zinc finger nuclease 3 |
-0.423 |
0.105 |
| 409 |
266097_at |
AT2G37970
|
AtHBP2, HBP2, haem-binding protein 2, SOUL-1 |
-0.422 |
0.052 |
| 410 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.421 |
-0.265 |
| 411 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.421 |
-0.144 |
| 412 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.420 |
-0.509 |
| 413 |
250960_at |
AT5G02940
|
unknown |
-0.418 |
-0.132 |
| 414 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
-0.417 |
-0.164 |
| 415 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.417 |
-0.093 |
| 416 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.417 |
0.247 |
| 417 |
247222_at |
AT5G64840
|
ABCF5, ATP-binding cassette F5, ATGCN5, general control non-repressible 5, GCN5, general control non-repressible 5 |
-0.416 |
-0.222 |
| 418 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
-0.416 |
-0.244 |
| 419 |
246792_at |
AT5G27290
|
unknown |
-0.415 |
-0.163 |
| 420 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.413 |
-0.300 |
| 421 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.413 |
-0.343 |
| 422 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.413 |
-0.089 |
| 423 |
254693_at |
AT4G17880
|
MYC4 |
-0.412 |
0.004 |
| 424 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.411 |
0.850 |
| 425 |
263632_at |
AT2G04795
|
unknown |
-0.409 |
0.059 |
| 426 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.409 |
-0.529 |
| 427 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.407 |
-0.722 |
| 428 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.404 |
-0.105 |
| 429 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.404 |
-0.035 |