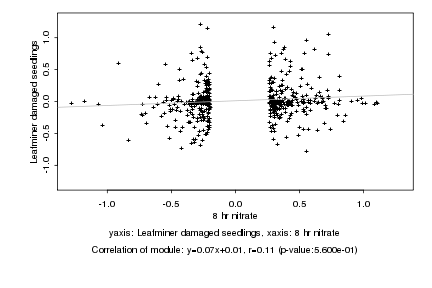
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
8 hr nitrate |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
250472_at |
AT5G10210
|
unknown |
1.103 |
-0.029 |
| 2 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
1.095 |
-0.011 |
| 3 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
1.085 |
-0.040 |
| 4 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
1.009 |
-0.020 |
| 5 |
252501_at |
AT3G46880
|
unknown |
0.987 |
-0.024 |
| 6 |
260624_at |
AT1G08100
|
ACH2, ATNRT2.2, NITRATE TRANSPORTER 2.2, NRT2.2, nitrate transporter 2.2, NRT2;2AT |
0.976 |
0.048 |
| 7 |
251908_at |
AT3G53770
|
[late embryogenesis abundant 3 (LEA3) family protein] |
0.947 |
0.023 |
| 8 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.901 |
0.014 |
| 9 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.854 |
-0.217 |
| 10 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.840 |
-0.305 |
| 11 |
262399_at |
AT1G49500
|
unknown |
0.812 |
0.187 |
| 12 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.811 |
0.029 |
| 13 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.805 |
0.405 |
| 14 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.801 |
-0.031 |
| 15 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.789 |
-0.216 |
| 16 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
0.767 |
-0.021 |
| 17 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.739 |
-0.425 |
| 18 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.726 |
1.055 |
| 19 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.726 |
0.744 |
| 20 |
252882_at |
AT4G39675
|
unknown |
0.724 |
0.080 |
| 21 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.722 |
0.060 |
| 22 |
247754_at |
AT5G59080
|
unknown |
0.715 |
0.178 |
| 23 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
0.704 |
-0.107 |
| 24 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.699 |
-0.062 |
| 25 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.698 |
-0.101 |
| 26 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.688 |
-0.339 |
| 27 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.680 |
0.014 |
| 28 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.677 |
0.071 |
| 29 |
253322_at |
AT4G33980
|
unknown |
0.676 |
0.060 |
| 30 |
252456_at |
AT3G47170
|
[HXXXD-type acyl-transferase family protein] |
0.673 |
-0.015 |
| 31 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.666 |
-0.012 |
| 32 |
260771_at |
AT1G49160
|
WNK7 |
0.657 |
0.093 |
| 33 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.654 |
0.386 |
| 34 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.654 |
0.150 |
| 35 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.644 |
-0.011 |
| 36 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.638 |
-0.453 |
| 37 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
0.638 |
-0.028 |
| 38 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.628 |
0.104 |
| 39 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
0.623 |
0.061 |
| 40 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.611 |
0.826 |
| 41 |
260750_at |
AT1G49100
|
[Leucine-rich repeat protein kinase family protein] |
0.610 |
0.006 |
| 42 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
0.589 |
0.213 |
| 43 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.585 |
-0.129 |
| 44 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.578 |
-0.019 |
| 45 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.578 |
0.028 |
| 46 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.569 |
-0.013 |
| 47 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.566 |
-0.434 |
| 48 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.565 |
0.019 |
| 49 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.556 |
0.273 |
| 50 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.554 |
-0.080 |
| 51 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.554 |
-0.772 |
| 52 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.552 |
0.971 |
| 53 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
0.550 |
-0.194 |
| 54 |
255945_at |
AT5G28610
|
unknown |
0.547 |
0.104 |
| 55 |
261996_at |
AT1G33830
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.537 |
-0.015 |
| 56 |
259996_at |
AT1G67910
|
unknown |
0.532 |
-0.150 |
| 57 |
262452_at |
AT1G11210
|
unknown |
0.532 |
0.762 |
| 58 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.531 |
0.037 |
| 59 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.528 |
-0.146 |
| 60 |
261684_at |
AT1G47400
|
unknown |
0.526 |
-0.436 |
| 61 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.525 |
-0.123 |
| 62 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.524 |
-0.001 |
| 63 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.524 |
-0.019 |
| 64 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.521 |
0.505 |
| 65 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.517 |
0.087 |
| 66 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.513 |
0.370 |
| 67 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.512 |
0.511 |
| 68 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.511 |
0.016 |
| 69 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.502 |
0.237 |
| 70 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
0.497 |
-0.403 |
| 71 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.488 |
-0.524 |
| 72 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
0.486 |
-0.058 |
| 73 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.481 |
-0.007 |
| 74 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.473 |
-0.144 |
| 75 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.473 |
0.126 |
| 76 |
249413_at |
AT5G39620
|
AtRABG1, RAB GTPase homolog G1, RABG1, RAB GTPase homolog G1 |
0.472 |
0.039 |
| 77 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.465 |
0.076 |
| 78 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
0.453 |
0.083 |
| 79 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.449 |
0.155 |
| 80 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.445 |
-0.051 |
| 81 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.444 |
0.103 |
| 82 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.441 |
0.029 |
| 83 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.434 |
0.201 |
| 84 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
0.432 |
-0.183 |
| 85 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
0.431 |
-0.003 |
| 86 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.431 |
-0.052 |
| 87 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.429 |
0.635 |
| 88 |
261921_at |
AT1G65900
|
unknown |
0.429 |
-0.205 |
| 89 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.425 |
0.549 |
| 90 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.422 |
-0.036 |
| 91 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.421 |
0.475 |
| 92 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.419 |
-0.257 |
| 93 |
253317_at |
AT4G33960
|
unknown |
0.418 |
-0.001 |
| 94 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.415 |
0.001 |
| 95 |
263213_at |
AT1G30560
|
AtG3Pp3, glycerol-3-phosphate permease 3, G3Pp3, glycerol-3-phosphate permease 3 |
0.415 |
-0.012 |
| 96 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.413 |
-0.062 |
| 97 |
245962_at |
AT5G19700
|
[MATE efflux family protein] |
0.413 |
0.002 |
| 98 |
259828_at |
AT1G72220
|
[RING/U-box superfamily protein] |
0.408 |
0.004 |
| 99 |
248356_at |
AT5G52350
|
ATEXO70A3, exocyst subunit exo70 family protein A3, EXO70A3, exocyst subunit exo70 family protein A3 |
0.408 |
-0.028 |
| 100 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
0.403 |
-0.012 |
| 101 |
265329_at |
AT2G18450
|
SDH1-2, succinate dehydrogenase 1-2 |
0.402 |
0.001 |
| 102 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.402 |
-0.311 |
| 103 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.402 |
0.221 |
| 104 |
249932_at |
AT5G22390
|
unknown |
0.400 |
-0.051 |
| 105 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.394 |
0.433 |
| 106 |
246054_at |
AT5G08360
|
unknown |
0.393 |
-0.042 |
| 107 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.392 |
-0.561 |
| 108 |
245353_at |
AT4G16000
|
unknown |
0.390 |
0.661 |
| 109 |
263556_at |
AT2G16365
|
[F-box family protein] |
0.388 |
-0.169 |
| 110 |
251560_at |
AT3G57920
|
SPL15, squamosa promoter binding protein-like 15 |
0.385 |
-0.104 |
| 111 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.385 |
0.198 |
| 112 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
0.384 |
0.063 |
| 113 |
260264_at |
AT1G68500
|
unknown |
0.381 |
0.057 |
| 114 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.377 |
0.856 |
| 115 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.376 |
-0.038 |
| 116 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.372 |
-0.096 |
| 117 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
0.371 |
-0.000 |
| 118 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.370 |
0.828 |
| 119 |
258275_at |
AT3G15760
|
unknown |
0.366 |
0.026 |
| 120 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.364 |
0.162 |
| 121 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.362 |
0.339 |
| 122 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.360 |
0.256 |
| 123 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.359 |
-0.190 |
| 124 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.358 |
-0.039 |
| 125 |
256370_at |
AT1G66780
|
[MATE efflux family protein] |
0.358 |
0.126 |
| 126 |
260754_at |
AT1G49000
|
unknown |
0.356 |
0.473 |
| 127 |
261791_at |
AT1G16170
|
unknown |
0.356 |
-0.070 |
| 128 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.355 |
0.266 |
| 129 |
260005_at |
AT1G67920
|
unknown |
0.354 |
0.768 |
| 130 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.351 |
-0.244 |
| 131 |
266222_at |
AT2G28780
|
unknown |
0.349 |
0.005 |
| 132 |
251380_at |
AT3G60700
|
unknown |
0.349 |
-0.060 |
| 133 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.345 |
-0.007 |
| 134 |
253671_at |
AT4G29990
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.344 |
-0.035 |
| 135 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.344 |
0.178 |
| 136 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.343 |
-0.242 |
| 137 |
262546_at |
AT1G31260
|
ZIP10, zinc transporter 10 precursor |
0.343 |
0.004 |
| 138 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.342 |
-0.035 |
| 139 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
0.342 |
-0.094 |
| 140 |
259015_at |
AT3G07350
|
unknown |
0.340 |
0.249 |
| 141 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.340 |
-0.118 |
| 142 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.340 |
0.078 |
| 143 |
267441_at |
AT2G19210
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.338 |
-0.018 |
| 144 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
0.337 |
-0.263 |
| 145 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.327 |
0.021 |
| 146 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.324 |
-0.112 |
| 147 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.323 |
-0.215 |
| 148 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
0.323 |
-0.101 |
| 149 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.322 |
-0.002 |
| 150 |
266176_at |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
0.322 |
-0.020 |
| 151 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.321 |
-0.666 |
| 152 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.320 |
-0.178 |
| 153 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.318 |
0.162 |
| 154 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.318 |
0.172 |
| 155 |
250855_at |
AT5G04730
|
[Ankyrin-repeat containing protein] |
0.315 |
-0.030 |
| 156 |
260325_at |
AT1G63940
|
MDAR6, monodehydroascorbate reductase 6 |
0.313 |
-0.079 |
| 157 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.313 |
0.113 |
| 158 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.313 |
-0.185 |
| 159 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
0.310 |
0.016 |
| 160 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.307 |
0.732 |
| 161 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.305 |
-0.348 |
| 162 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.304 |
0.939 |
| 163 |
255456_at |
AT4G02920
|
unknown |
0.304 |
-0.209 |
| 164 |
260688_at |
AT1G17665
|
unknown |
0.303 |
0.022 |
| 165 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.303 |
-0.026 |
| 166 |
250463_at |
AT5G10030
|
OBF4, OCS ELEMENT BINDING FACTOR 4, TGA4, TGACG motif-binding factor 4 |
0.303 |
-0.105 |
| 167 |
255377_at |
AT4G03500
|
[Ankyrin repeat family protein] |
0.303 |
-0.135 |
| 168 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.302 |
-0.467 |
| 169 |
250943_at |
AT5G03360
|
[DC1 domain-containing protein] |
0.302 |
-0.066 |
| 170 |
265387_at |
AT2G20670
|
unknown |
0.302 |
-0.242 |
| 171 |
255041_at |
AT4G09620
|
[Mitochondrial transcription termination factor family protein] |
0.302 |
-0.146 |
| 172 |
245977_at |
AT5G13110
|
G6PD2, glucose-6-phosphate dehydrogenase 2 |
0.300 |
-0.112 |
| 173 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
0.299 |
-0.014 |
| 174 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.299 |
0.370 |
| 175 |
261980_at |
AT1G33820
|
unknown |
0.298 |
-0.011 |
| 176 |
248898_at |
AT5G46370
|
ATKCO2, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 2, ATTPK2, TANDEM PORE K+ CHANNEL 2, KCO2, Ca2+ activated outward rectifying K+ channel 2 |
0.298 |
-0.035 |
| 177 |
261543_at |
AT1G63550
|
[Receptor-like protein kinase-related family protein] |
0.297 |
-0.008 |
| 178 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.297 |
-0.054 |
| 179 |
250559_at |
AT5G08010
|
unknown |
0.295 |
-0.161 |
| 180 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.295 |
-0.015 |
| 181 |
263182_at |
AT1G05575
|
unknown |
0.294 |
1.162 |
| 182 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.293 |
0.110 |
| 183 |
265543_at |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.293 |
-0.014 |
| 184 |
255940_at |
AT1G20380
|
[Prolyl oligopeptidase family protein] |
0.293 |
-0.591 |
| 185 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.293 |
-0.414 |
| 186 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.290 |
0.304 |
| 187 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
0.290 |
-0.121 |
| 188 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
0.290 |
-0.084 |
| 189 |
263919_at |
AT2G36470
|
unknown |
0.289 |
0.063 |
| 190 |
261487_at |
AT1G14340
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.288 |
-0.002 |
| 191 |
250828_at |
AT5G05250
|
unknown |
0.287 |
-0.367 |
| 192 |
263572_at |
AT2G17060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.286 |
0.043 |
| 193 |
250223_at |
AT5G14070
|
ROXY2 |
0.286 |
-0.033 |
| 194 |
257530_at |
AT3G03040
|
[F-box/RNI-like superfamily protein] |
0.284 |
-0.093 |
| 195 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.284 |
0.301 |
| 196 |
252293_at |
AT3G48990
|
AAE3, ACYL-ACTIVATING ENZYME 3 |
0.284 |
0.068 |
| 197 |
257550_at |
AT3G18460
|
[PLAC8 family protein] |
0.283 |
0.006 |
| 198 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.282 |
0.181 |
| 199 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
0.281 |
-0.013 |
| 200 |
257466_at |
AT1G62840
|
unknown |
0.281 |
0.680 |
| 201 |
250594_at |
AT5G07760
|
[formin homology 2 domain-containing protein / FH2 domain-containing protein] |
0.280 |
0.014 |
| 202 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.278 |
-0.076 |
| 203 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
0.276 |
-0.026 |
| 204 |
258939_at |
AT3G10020
|
unknown |
0.276 |
0.296 |
| 205 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
0.275 |
-0.061 |
| 206 |
256799_at |
AT3G18560
|
unknown |
0.275 |
0.390 |
| 207 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
0.274 |
-0.468 |
| 208 |
253989_at |
AT4G26130
|
unknown |
0.274 |
0.190 |
| 209 |
248416_at |
AT5G51630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.273 |
-0.128 |
| 210 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
0.273 |
0.225 |
| 211 |
246142_at |
AT5G19970
|
unknown |
0.272 |
-0.183 |
| 212 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
0.271 |
-0.117 |
| 213 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.271 |
0.094 |
| 214 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.270 |
-0.224 |
| 215 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.270 |
0.048 |
| 216 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.269 |
0.165 |
| 217 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.268 |
-0.013 |
| 218 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.268 |
0.365 |
| 219 |
248267_at |
AT5G53460
|
GLT1, NADH-dependent glutamate synthase 1 |
0.267 |
0.039 |
| 220 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.266 |
0.767 |
| 221 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
0.266 |
-0.401 |
| 222 |
261193_at |
AT1G32920
|
unknown |
0.265 |
0.578 |
| 223 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.264 |
-0.005 |
| 224 |
267032_at |
AT2G38490
|
CIPK22, CBL-interacting protein kinase 22, SnRK3.19, SNF1-RELATED PROTEIN KINASE 3.19 |
0.264 |
0.081 |
| 225 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
0.264 |
-0.328 |
| 226 |
262885_at |
AT1G64740
|
TUA1, alpha-1 tubulin |
0.263 |
-0.023 |
| 227 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.263 |
0.332 |
| 228 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.261 |
0.639 |
| 229 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.261 |
0.160 |
| 230 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.261 |
0.297 |
| 231 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-1.285 |
-0.016 |
| 232 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-1.181 |
0.016 |
| 233 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-1.076 |
-0.043 |
| 234 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-1.042 |
-0.373 |
| 235 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.917 |
0.609 |
| 236 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.842 |
-0.597 |
| 237 |
255957_at |
AT1G22160
|
unknown |
-0.735 |
-0.196 |
| 238 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.733 |
-0.207 |
| 239 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.727 |
-0.034 |
| 240 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.704 |
-0.178 |
| 241 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
-0.696 |
-0.342 |
| 242 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
-0.678 |
0.069 |
| 243 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.644 |
-0.088 |
| 244 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
-0.627 |
0.068 |
| 245 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.613 |
-0.040 |
| 246 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
-0.606 |
0.279 |
| 247 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.578 |
-0.234 |
| 248 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.562 |
-0.009 |
| 249 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.558 |
-0.179 |
| 250 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.546 |
0.595 |
| 251 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.542 |
0.074 |
| 252 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.539 |
0.028 |
| 253 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.536 |
-0.391 |
| 254 |
247797_at |
AT5G58780
|
AtcPT5, AtHEPS, cPT5, cis -prenyltransferase 5, HEPS, heptaprenyl diphosphate synthase |
-0.520 |
-0.050 |
| 255 |
262811_at |
AT1G11700
|
unknown |
-0.519 |
-0.572 |
| 256 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.513 |
-0.144 |
| 257 |
266564_at |
AT2G24000
|
scpl22, serine carboxypeptidase-like 22 |
-0.507 |
-0.124 |
| 258 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.503 |
0.022 |
| 259 |
266565_at |
AT2G24010
|
scpl23, serine carboxypeptidase-like 23 |
-0.492 |
-0.124 |
| 260 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.491 |
0.043 |
| 261 |
260535_at |
AT2G43390
|
unknown |
-0.490 |
0.034 |
| 262 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
-0.487 |
0.051 |
| 263 |
262373_at |
AT1G73120
|
unknown |
-0.478 |
-0.043 |
| 264 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.470 |
-0.398 |
| 265 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.470 |
-0.287 |
| 266 |
265569_at |
AT2G05620
|
AtPGR5, PGR5, proton gradient regulation 5 |
-0.464 |
-0.127 |
| 267 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.459 |
-0.090 |
| 268 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
-0.455 |
0.002 |
| 269 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.450 |
0.086 |
| 270 |
264842_at |
AT1G03700
|
[Uncharacterised protein family (UPF0497)] |
-0.448 |
0.047 |
| 271 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
-0.442 |
0.512 |
| 272 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.439 |
0.330 |
| 273 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-0.430 |
-0.145 |
| 274 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.429 |
-0.461 |
| 275 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
-0.425 |
0.011 |
| 276 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.423 |
-0.730 |
| 277 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.418 |
-0.402 |
| 278 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
-0.409 |
0.355 |
| 279 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.403 |
-0.042 |
| 280 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.383 |
-0.008 |
| 281 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.377 |
-0.126 |
| 282 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.376 |
-0.213 |
| 283 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.371 |
-0.005 |
| 284 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.368 |
-0.327 |
| 285 |
260298_at |
AT1G80320
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.364 |
0.013 |
| 286 |
249622_at |
AT5G37550
|
unknown |
-0.364 |
-0.044 |
| 287 |
247751_at |
AT5G59050
|
unknown |
-0.363 |
-0.032 |
| 288 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.361 |
-0.319 |
| 289 |
245401_at |
AT4G17670
|
unknown |
-0.355 |
-0.171 |
| 290 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.352 |
-0.327 |
| 291 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.348 |
-0.650 |
| 292 |
265769_at |
AT2G48090
|
unknown |
-0.347 |
-0.025 |
| 293 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.345 |
0.762 |
| 294 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.345 |
-0.039 |
| 295 |
249010_at |
AT5G44580
|
unknown |
-0.344 |
-0.018 |
| 296 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.344 |
-0.073 |
| 297 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.340 |
0.047 |
| 298 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.339 |
-0.016 |
| 299 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.339 |
-0.125 |
| 300 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.338 |
-0.203 |
| 301 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
-0.338 |
0.116 |
| 302 |
259991_at |
AT1G68040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.337 |
0.012 |
| 303 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.337 |
0.650 |
| 304 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.332 |
0.114 |
| 305 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.332 |
-0.583 |
| 306 |
258854_at |
AT3G02100
|
[UDP-Glycosyltransferase superfamily protein] |
-0.328 |
-0.022 |
| 307 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.328 |
-0.241 |
| 308 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.325 |
-0.396 |
| 309 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.323 |
-0.627 |
| 310 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
-0.323 |
-0.069 |
| 311 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.318 |
-0.586 |
| 312 |
263652_at |
AT1G04330
|
unknown |
-0.316 |
-0.007 |
| 313 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.313 |
-0.021 |
| 314 |
251079_at |
AT5G02000
|
unknown |
-0.310 |
-0.033 |
| 315 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.307 |
0.314 |
| 316 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.306 |
-0.013 |
| 317 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.302 |
-0.265 |
| 318 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.302 |
0.033 |
| 319 |
265025_at |
AT1G24575
|
unknown |
-0.300 |
0.240 |
| 320 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.298 |
-0.303 |
| 321 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.297 |
0.676 |
| 322 |
247992_at |
AT5G56520
|
unknown |
-0.296 |
0.022 |
| 323 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.295 |
0.311 |
| 324 |
245663_at |
AT1G28170
|
SOT7, sulphotransferase 7 |
-0.294 |
-0.028 |
| 325 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.294 |
0.009 |
| 326 |
265539_at |
AT2G15830
|
unknown |
-0.292 |
0.140 |
| 327 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.292 |
-0.522 |
| 328 |
257348_at |
AT2G42140
|
[VQ motif-containing protein] |
-0.288 |
0.067 |
| 329 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.287 |
-0.193 |
| 330 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.286 |
-0.106 |
| 331 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.284 |
-0.015 |
| 332 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.281 |
-0.466 |
| 333 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.280 |
0.038 |
| 334 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
-0.280 |
-0.250 |
| 335 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.278 |
0.852 |
| 336 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.278 |
0.435 |
| 337 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.278 |
-0.135 |
| 338 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.277 |
0.044 |
| 339 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
-0.275 |
0.050 |
| 340 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.275 |
1.214 |
| 341 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.275 |
-0.687 |
| 342 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.274 |
-0.019 |
| 343 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.273 |
0.091 |
| 344 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.273 |
-0.027 |
| 345 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.272 |
0.786 |
| 346 |
256926_at |
AT3G22540
|
unknown |
-0.272 |
-0.100 |
| 347 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.272 |
-0.284 |
| 348 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.272 |
0.024 |
| 349 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
-0.272 |
-0.022 |
| 350 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.271 |
-0.138 |
| 351 |
264621_at |
AT2G17700
|
STY8, serine/threonine/tyrosine kinase 8 |
-0.270 |
0.062 |
| 352 |
264082_at |
AT2G28570
|
unknown |
-0.269 |
0.038 |
| 353 |
266597_at |
AT2G46130
|
ATWRKY43, WRKY DNA-BINDING PROTEIN 43, WRKY43, WRKY DNA-binding protein 43 |
-0.268 |
0.015 |
| 354 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
-0.268 |
0.443 |
| 355 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.267 |
0.082 |
| 356 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
-0.265 |
-0.031 |
| 357 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
-0.264 |
-0.188 |
| 358 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.264 |
-0.168 |
| 359 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
-0.264 |
-0.001 |
| 360 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.262 |
0.778 |
| 361 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.262 |
0.061 |
| 362 |
245692_at |
AT5G04150
|
BHLH101 |
-0.261 |
-0.605 |
| 363 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
-0.261 |
0.046 |
| 364 |
260147_at |
AT1G52790
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.259 |
0.040 |
| 365 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.257 |
-0.245 |
| 366 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.257 |
-0.228 |
| 367 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.255 |
0.086 |
| 368 |
262598_at |
AT1G15260
|
unknown |
-0.255 |
-0.442 |
| 369 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.254 |
-0.023 |
| 370 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.253 |
-0.287 |
| 371 |
246477_at |
AT5G16770
|
AtMYB9, myb domain protein 9, MYB9, myb domain protein 9 |
-0.251 |
-0.042 |
| 372 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.250 |
-0.016 |
| 373 |
249922_at |
AT5G19140
|
AILP1, ATAILP1 |
-0.250 |
0.146 |
| 374 |
252017_at |
AT3G52970
|
CYP76G1, cytochrome P450, family 76, subfamily G, polypeptide 1 |
-0.250 |
-0.039 |
| 375 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.248 |
0.048 |
| 376 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.247 |
0.594 |
| 377 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.246 |
0.338 |
| 378 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.246 |
-0.252 |
| 379 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
-0.243 |
-0.143 |
| 380 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
-0.241 |
0.067 |
| 381 |
247769_at |
AT5G58910
|
LAC16, laccase 16 |
-0.240 |
0.010 |
| 382 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
-0.239 |
0.050 |
| 383 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.237 |
-0.505 |
| 384 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.237 |
-0.035 |
| 385 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.237 |
0.025 |
| 386 |
266835_at |
AT2G29990
|
NDA2, alternative NAD(P)H dehydrogenase 2 |
-0.236 |
0.165 |
| 387 |
246584_at |
AT5G14730
|
unknown |
-0.235 |
0.298 |
| 388 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.234 |
0.024 |
| 389 |
267133_at |
AT2G23440
|
unknown |
-0.232 |
-0.008 |
| 390 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.232 |
0.217 |
| 391 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
-0.231 |
0.401 |
| 392 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.230 |
0.256 |
| 393 |
245056_at |
AT2G26480
|
UGT76D1, UDP-glucosyl transferase 76D1 |
-0.230 |
0.050 |
| 394 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.230 |
0.168 |
| 395 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.230 |
0.544 |
| 396 |
247020_at |
AT5G67020
|
unknown |
-0.229 |
0.156 |
| 397 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.228 |
-0.503 |
| 398 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.227 |
-0.008 |
| 399 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
-0.227 |
0.065 |
| 400 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.227 |
0.034 |
| 401 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.226 |
-0.121 |
| 402 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-0.226 |
0.186 |
| 403 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
-0.226 |
0.315 |
| 404 |
246067_at |
AT5G19410
|
ABCG23, ATP-binding cassette G23 |
-0.225 |
-0.003 |
| 405 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.225 |
-0.010 |
| 406 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
-0.224 |
0.049 |
| 407 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
-0.224 |
-0.303 |
| 408 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.224 |
-0.475 |
| 409 |
267319_at |
AT2G34660
|
ABCC2, ATP-binding cassette C2, AtABCC2, Arabidopsis thaliana ATP-binding cassette C2, ATMRP2, multidrug resistance-associated protein 2, EST4, MRP2, multidrug resistance-associated protein 2 |
-0.223 |
-0.230 |
| 410 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.223 |
0.293 |
| 411 |
247231_at |
AT5G65230
|
AtMYB53, myb domain protein 53, MYB53, myb domain protein 53 |
-0.223 |
-0.026 |
| 412 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.223 |
-0.025 |
| 413 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.223 |
-0.040 |
| 414 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.223 |
0.096 |
| 415 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.222 |
-0.385 |
| 416 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.222 |
-0.062 |
| 417 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.222 |
1.146 |
| 418 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
-0.220 |
0.193 |
| 419 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.219 |
0.315 |
| 420 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.219 |
0.693 |
| 421 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
-0.218 |
0.183 |
| 422 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.218 |
0.005 |
| 423 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
-0.218 |
-0.019 |
| 424 |
261292_at |
AT1G36940
|
unknown |
-0.217 |
0.013 |
| 425 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
-0.217 |
-0.121 |
| 426 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.216 |
0.153 |
| 427 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
-0.215 |
0.000 |
| 428 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.214 |
0.260 |
| 429 |
261089_at |
AT1G07570
|
APK1, APK1A |
-0.214 |
-0.295 |
| 430 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
-0.213 |
0.058 |
| 431 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.213 |
-0.176 |
| 432 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.213 |
-0.162 |
| 433 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.212 |
-0.299 |
| 434 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
-0.212 |
0.354 |
| 435 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.211 |
-0.435 |
| 436 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.209 |
-0.263 |
| 437 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
-0.209 |
-0.121 |
| 438 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.209 |
-0.040 |
| 439 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
-0.208 |
0.442 |
| 440 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.208 |
-0.374 |
| 441 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.207 |
-0.045 |
| 442 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.206 |
0.036 |
| 443 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.206 |
-0.064 |
| 444 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.206 |
-0.030 |
| 445 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.206 |
0.214 |
| 446 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.205 |
-0.401 |
| 447 |
248697_at |
AT5G48370
|
[Thioesterase/thiol ester dehydrase-isomerase superfamily protein] |
-0.205 |
0.089 |
| 448 |
253022_at |
AT4G38060
|
CCI2, Clavata complex interactor 2 |
-0.205 |
-0.102 |
| 449 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.205 |
-0.065 |
| 450 |
262162_at |
AT1G78020
|
unknown |
-0.204 |
-0.322 |
| 451 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.204 |
0.201 |
| 452 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
-0.204 |
0.018 |
| 453 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
-0.204 |
0.136 |
| 454 |
251931_at |
AT3G53950
|
[glyoxal oxidase-related protein] |
-0.203 |
-0.256 |
| 455 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.203 |
0.279 |
| 456 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
-0.203 |
0.310 |
| 457 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.203 |
-0.044 |
| 458 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
-0.203 |
0.009 |
| 459 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.202 |
-0.079 |