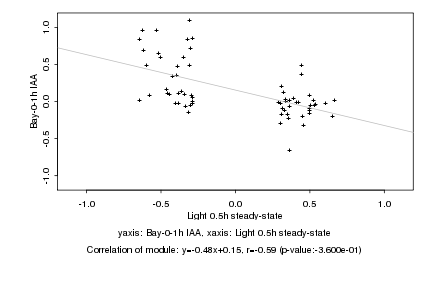
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Light 0.5h steady-state |
Log2 signal ratio
Bay-0-1h IAA |
| 1 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.659 |
0.021 |
| 2 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.646 |
-0.198 |
| 3 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
0.600 |
-0.022 |
| 4 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.534 |
-0.038 |
| 5 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.528 |
-0.044 |
| 6 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.524 |
0.017 |
| 7 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.503 |
-0.051 |
| 8 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.497 |
-0.161 |
| 9 |
256518_at |
AT1G66080
|
unknown |
0.492 |
-0.108 |
| 10 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.491 |
-0.088 |
| 11 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.491 |
0.093 |
| 12 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.453 |
-0.311 |
| 13 |
253859_at |
AT4G27657
|
unknown |
0.448 |
-0.198 |
| 14 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.443 |
0.369 |
| 15 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.442 |
0.495 |
| 16 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.420 |
-0.002 |
| 17 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.405 |
-0.012 |
| 18 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.384 |
0.054 |
| 19 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.360 |
0.023 |
| 20 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.358 |
-0.650 |
| 21 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.357 |
-0.056 |
| 22 |
257925_at |
AT3G23170
|
unknown |
0.353 |
-0.222 |
| 23 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.348 |
-0.168 |
| 24 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.333 |
0.001 |
| 25 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.333 |
0.032 |
| 26 |
251727_at |
AT3G56290
|
unknown |
0.332 |
0.010 |
| 27 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.326 |
-0.117 |
| 28 |
256097_at |
AT1G13670
|
unknown |
0.319 |
0.132 |
| 29 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.313 |
-0.092 |
| 30 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.303 |
0.204 |
| 31 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.303 |
-0.166 |
| 32 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.300 |
-0.017 |
| 33 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.298 |
-0.287 |
| 34 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.289 |
-0.011 |
| 35 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.647 |
0.845 |
| 36 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.645 |
0.023 |
| 37 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.629 |
0.968 |
| 38 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.624 |
0.697 |
| 39 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.601 |
0.486 |
| 40 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.582 |
0.082 |
| 41 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.533 |
0.959 |
| 42 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.518 |
0.649 |
| 43 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.505 |
0.605 |
| 44 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.464 |
0.173 |
| 45 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.459 |
0.117 |
| 46 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.443 |
0.103 |
| 47 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.423 |
0.342 |
| 48 |
258935_at |
AT3G10120
|
unknown |
-0.406 |
-0.024 |
| 49 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.396 |
0.358 |
| 50 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.395 |
0.473 |
| 51 |
263151_at |
AT1G54120
|
unknown |
-0.388 |
0.109 |
| 52 |
251058_at |
AT5G01790
|
unknown |
-0.386 |
-0.024 |
| 53 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.367 |
0.138 |
| 54 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.354 |
0.599 |
| 55 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.344 |
0.105 |
| 56 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.339 |
-0.054 |
| 57 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.327 |
0.839 |
| 58 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.320 |
-0.135 |
| 59 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.312 |
0.490 |
| 60 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.311 |
1.106 |
| 61 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
-0.308 |
-0.042 |
| 62 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.308 |
0.720 |
| 63 |
247457_at |
AT5G62170
|
TRM25, TON1 Recruiting Motif 25 |
-0.296 |
0.088 |
| 64 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.295 |
0.860 |
| 65 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.295 |
-0.014 |
| 66 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.293 |
0.013 |
| 67 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.292 |
0.063 |