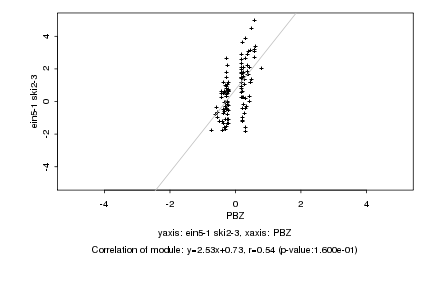
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PBZ |
Log2 signal ratio
ein5-1 ski2-3 |
| 1 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.780 |
2.045 |
| 2 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.600 |
3.379 |
| 3 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.579 |
3.118 |
| 4 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.578 |
5.030 |
| 5 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.575 |
2.745 |
| 6 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.559 |
3.214 |
| 7 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.474 |
1.384 |
| 8 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.465 |
4.488 |
| 9 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.451 |
1.181 |
| 10 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.443 |
3.178 |
| 11 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.430 |
1.172 |
| 12 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.426 |
0.362 |
| 13 |
257798_at |
AT3G15950
|
NAI2 |
0.410 |
0.031 |
| 14 |
249454_at |
AT5G39520
|
unknown |
0.402 |
2.094 |
| 15 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.378 |
1.675 |
| 16 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.375 |
3.115 |
| 17 |
256924_at |
AT3G29590
|
AT5MAT |
0.365 |
2.910 |
| 18 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.358 |
1.675 |
| 19 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.353 |
2.266 |
| 20 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.352 |
1.863 |
| 21 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.309 |
-0.293 |
| 22 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.303 |
0.239 |
| 23 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.300 |
3.866 |
| 24 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.299 |
-1.815 |
| 25 |
254391_at |
AT4G21590
|
ENDO3, endonuclease 3 |
0.299 |
-0.406 |
| 26 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.290 |
-1.574 |
| 27 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.279 |
2.643 |
| 28 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.269 |
1.369 |
| 29 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.259 |
-0.718 |
| 30 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.257 |
1.104 |
| 31 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
0.252 |
1.372 |
| 32 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.235 |
1.790 |
| 33 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.231 |
1.547 |
| 34 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.219 |
-0.181 |
| 35 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
0.219 |
0.308 |
| 36 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.216 |
2.106 |
| 37 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.210 |
1.774 |
| 38 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
0.210 |
-1.178 |
| 39 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.206 |
0.247 |
| 40 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.203 |
0.676 |
| 41 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.203 |
-0.406 |
| 42 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
0.198 |
-0.949 |
| 43 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.190 |
3.650 |
| 44 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
0.186 |
-1.116 |
| 45 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.185 |
1.486 |
| 46 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.181 |
0.817 |
| 47 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.179 |
1.759 |
| 48 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.178 |
0.254 |
| 49 |
256883_at |
AT3G26440
|
unknown |
0.177 |
0.923 |
| 50 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.176 |
2.128 |
| 51 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.174 |
1.826 |
| 52 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.174 |
2.907 |
| 53 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.171 |
0.591 |
| 54 |
255900_at |
AT1G17830
|
unknown |
0.170 |
2.359 |
| 55 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.169 |
1.197 |
| 56 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.169 |
1.464 |
| 57 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.165 |
1.522 |
| 58 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.165 |
2.018 |
| 59 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.161 |
2.581 |
| 60 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.751 |
-1.733 |
| 61 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.622 |
-0.737 |
| 62 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.593 |
-0.355 |
| 63 |
254574_at |
AT4G19430
|
unknown |
-0.565 |
-0.967 |
| 64 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.561 |
-0.642 |
| 65 |
247878_at |
AT5G57760
|
unknown |
-0.491 |
-1.179 |
| 66 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.446 |
0.256 |
| 67 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.438 |
0.671 |
| 68 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.431 |
0.510 |
| 69 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.418 |
-1.740 |
| 70 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.416 |
-1.179 |
| 71 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.392 |
-0.436 |
| 72 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.388 |
-1.336 |
| 73 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.385 |
1.197 |
| 74 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.380 |
-0.674 |
| 75 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.372 |
0.524 |
| 76 |
256066_at |
AT1G06980
|
unknown |
-0.361 |
-0.038 |
| 77 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.347 |
0.588 |
| 78 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.344 |
-1.583 |
| 79 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.336 |
0.655 |
| 80 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.335 |
-0.600 |
| 81 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.332 |
-0.038 |
| 82 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.328 |
-1.658 |
| 83 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.326 |
-0.260 |
| 84 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.322 |
0.984 |
| 85 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.307 |
-1.087 |
| 86 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.302 |
-0.406 |
| 87 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.300 |
0.953 |
| 88 |
250936_at |
AT5G03120
|
unknown |
-0.299 |
0.348 |
| 89 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.297 |
2.693 |
| 90 |
267344_at |
AT2G44230
|
unknown |
-0.296 |
0.431 |
| 91 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.295 |
-0.406 |
| 92 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.285 |
1.501 |
| 93 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.281 |
1.813 |
| 94 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.280 |
-0.483 |
| 95 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.279 |
-1.509 |
| 96 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.278 |
0.339 |
| 97 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.277 |
0.856 |
| 98 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.271 |
-0.166 |
| 99 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.268 |
-0.450 |
| 100 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.264 |
-0.787 |
| 101 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.264 |
0.696 |
| 102 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.263 |
1.085 |
| 103 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.262 |
0.500 |
| 104 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.261 |
0.055 |
| 105 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.256 |
-1.336 |
| 106 |
252073_at |
AT3G51750
|
unknown |
-0.256 |
-1.087 |
| 107 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.256 |
-0.031 |
| 108 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.247 |
2.216 |
| 109 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.240 |
0.712 |
| 110 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.235 |
-1.084 |
| 111 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.224 |
-1.336 |
| 112 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.224 |
-0.541 |
| 113 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.221 |
0.769 |
| 114 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.219 |
-0.243 |
| 115 |
263688_at |
AT1G26920
|
unknown |
-0.218 |
0.658 |
| 116 |
257502_at |
AT1G78110
|
unknown |
-0.217 |
1.201 |
| 117 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.216 |
0.633 |