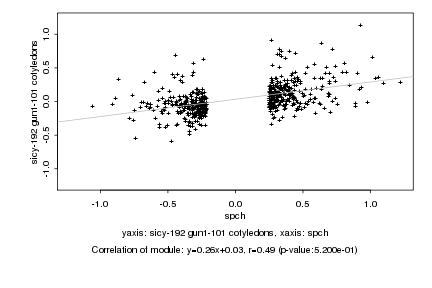
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
spch |
Log2 signal ratio
sicy-192 gun1-101 cotyledons |
| 1 |
260668_at |
AT1G19530
|
unknown |
1.219 |
0.297 |
| 2 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.095 |
0.274 |
| 3 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.057 |
0.358 |
| 4 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
1.034 |
0.349 |
| 5 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
1.008 |
0.662 |
| 6 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.976 |
-0.003 |
| 7 |
258327_at |
AT3G22640
|
PAP85 |
0.932 |
0.221 |
| 8 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.925 |
1.133 |
| 9 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.915 |
0.183 |
| 10 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.900 |
0.430 |
| 11 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.885 |
-0.028 |
| 12 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.884 |
-0.060 |
| 13 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.840 |
0.251 |
| 14 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.820 |
0.437 |
| 15 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.802 |
0.569 |
| 16 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.790 |
0.437 |
| 17 |
257670_at |
AT3G20340
|
unknown |
0.752 |
-0.040 |
| 18 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.739 |
0.057 |
| 19 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.738 |
0.529 |
| 20 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.736 |
0.307 |
| 21 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.725 |
0.365 |
| 22 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.716 |
0.778 |
| 23 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.712 |
0.012 |
| 24 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.700 |
-0.151 |
| 25 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.695 |
0.113 |
| 26 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.694 |
0.276 |
| 27 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.691 |
0.428 |
| 28 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.689 |
0.310 |
| 29 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.684 |
0.134 |
| 30 |
263475_at |
AT2G31945
|
unknown |
0.683 |
0.075 |
| 31 |
264580_at |
AT1G05340
|
unknown |
0.668 |
0.516 |
| 32 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.660 |
0.431 |
| 33 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.655 |
-0.092 |
| 34 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.643 |
0.350 |
| 35 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.641 |
0.225 |
| 36 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.634 |
0.193 |
| 37 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.631 |
0.870 |
| 38 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.626 |
0.344 |
| 39 |
259982_at |
AT1G76410
|
ATL8 |
0.623 |
-0.037 |
| 40 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.621 |
-0.021 |
| 41 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.598 |
0.200 |
| 42 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.587 |
-0.174 |
| 43 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.587 |
0.050 |
| 44 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.585 |
0.349 |
| 45 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.585 |
0.563 |
| 46 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.578 |
0.049 |
| 47 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.571 |
-0.013 |
| 48 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.559 |
0.191 |
| 49 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.552 |
0.112 |
| 50 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.550 |
0.038 |
| 51 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.536 |
0.014 |
| 52 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.530 |
-0.038 |
| 53 |
264774_at |
AT1G22890
|
unknown |
0.530 |
0.506 |
| 54 |
252882_at |
AT4G39675
|
unknown |
0.517 |
0.184 |
| 55 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.515 |
-0.020 |
| 56 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.514 |
0.424 |
| 57 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.507 |
-0.075 |
| 58 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.505 |
0.100 |
| 59 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.498 |
0.089 |
| 60 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.496 |
0.120 |
| 61 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.484 |
-0.117 |
| 62 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.479 |
0.298 |
| 63 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.479 |
0.268 |
| 64 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.478 |
-0.021 |
| 65 |
263118_at |
AT1G03090
|
MCCA |
0.476 |
-0.019 |
| 66 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.474 |
0.154 |
| 67 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.467 |
-0.061 |
| 68 |
262033_at |
AT1G37140
|
MCT1, MEI2 C-terminal RRM only like 1 |
0.462 |
-0.113 |
| 69 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.461 |
0.340 |
| 70 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.457 |
-0.000 |
| 71 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.457 |
-0.094 |
| 72 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.455 |
0.302 |
| 73 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.453 |
0.361 |
| 74 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.452 |
-0.125 |
| 75 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.447 |
0.024 |
| 76 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.446 |
0.251 |
| 77 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.444 |
0.724 |
| 78 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.442 |
0.300 |
| 79 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.440 |
0.435 |
| 80 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.437 |
0.239 |
| 81 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.437 |
-0.213 |
| 82 |
255285_at |
AT4G04630
|
unknown |
0.436 |
-0.030 |
| 83 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.436 |
0.057 |
| 84 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.435 |
0.352 |
| 85 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.433 |
0.166 |
| 86 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.430 |
0.070 |
| 87 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.429 |
0.284 |
| 88 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.424 |
-0.018 |
| 89 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.423 |
0.034 |
| 90 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.423 |
0.085 |
| 91 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.421 |
-0.054 |
| 92 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.418 |
0.061 |
| 93 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.416 |
0.422 |
| 94 |
261567_at |
AT1G33055
|
unknown |
0.415 |
0.124 |
| 95 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.414 |
0.198 |
| 96 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.414 |
0.319 |
| 97 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.412 |
0.134 |
| 98 |
245352_at |
AT4G15490
|
UGT84A3 |
0.411 |
0.389 |
| 99 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
0.402 |
0.096 |
| 100 |
264590_at |
AT2G17710
|
unknown |
0.402 |
0.102 |
| 101 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.398 |
0.046 |
| 102 |
262607_at |
AT1G13990
|
unknown |
0.396 |
0.222 |
| 103 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.395 |
0.006 |
| 104 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.395 |
0.140 |
| 105 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.395 |
0.753 |
| 106 |
258275_at |
AT3G15760
|
unknown |
0.393 |
0.101 |
| 107 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.392 |
0.190 |
| 108 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.392 |
0.115 |
| 109 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.390 |
0.170 |
| 110 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.387 |
0.163 |
| 111 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.386 |
0.164 |
| 112 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.385 |
0.038 |
| 113 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.383 |
0.282 |
| 114 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.381 |
0.237 |
| 115 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.380 |
0.278 |
| 116 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.379 |
0.021 |
| 117 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.379 |
0.321 |
| 118 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.377 |
-0.031 |
| 119 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.373 |
0.009 |
| 120 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.373 |
0.164 |
| 121 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.370 |
0.192 |
| 122 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.367 |
0.242 |
| 123 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.367 |
0.090 |
| 124 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.366 |
0.255 |
| 125 |
254638_at |
AT4G18740
|
[Rho termination factor] |
0.366 |
-0.093 |
| 126 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.364 |
0.648 |
| 127 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.364 |
-0.044 |
| 128 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.364 |
0.098 |
| 129 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.361 |
0.214 |
| 130 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.361 |
0.325 |
| 131 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.360 |
-0.115 |
| 132 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.358 |
0.129 |
| 133 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.356 |
-0.070 |
| 134 |
263570_at |
AT2G27150
|
AAO3, abscisic aldehyde oxidase 3, AOdelta, Aldehyde oxidase delta, At-AO3, Arabidopsis thaliana aldehyde oxidase 3, AtAAO3 |
0.351 |
0.126 |
| 135 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.351 |
-0.045 |
| 136 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.351 |
0.034 |
| 137 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.350 |
-0.056 |
| 138 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.349 |
0.222 |
| 139 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.347 |
0.002 |
| 140 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.344 |
0.012 |
| 141 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.344 |
0.017 |
| 142 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.342 |
-0.225 |
| 143 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.341 |
0.176 |
| 144 |
264238_at |
AT1G54740
|
unknown |
0.340 |
0.029 |
| 145 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.340 |
0.758 |
| 146 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.337 |
0.207 |
| 147 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.337 |
0.212 |
| 148 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.336 |
0.073 |
| 149 |
258262_at |
AT3G15770
|
unknown |
0.336 |
0.221 |
| 150 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.335 |
0.677 |
| 151 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.334 |
0.382 |
| 152 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.334 |
0.070 |
| 153 |
250098_at |
AT5G17350
|
unknown |
0.330 |
0.111 |
| 154 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.330 |
0.294 |
| 155 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.329 |
0.179 |
| 156 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.328 |
-0.005 |
| 157 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.328 |
0.247 |
| 158 |
261558_at |
AT1G01770
|
unknown |
0.327 |
0.089 |
| 159 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.326 |
0.511 |
| 160 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.325 |
-0.039 |
| 161 |
261205_at |
AT1G12790
|
unknown |
0.324 |
0.143 |
| 162 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.324 |
-0.271 |
| 163 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.323 |
0.707 |
| 164 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.323 |
0.086 |
| 165 |
252539_at |
AT3G45730
|
unknown |
0.322 |
0.781 |
| 166 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.322 |
0.121 |
| 167 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.321 |
0.088 |
| 168 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.319 |
0.196 |
| 169 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.318 |
0.077 |
| 170 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.316 |
-0.045 |
| 171 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.315 |
0.146 |
| 172 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.315 |
0.019 |
| 173 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.313 |
0.212 |
| 174 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.311 |
0.298 |
| 175 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.311 |
0.061 |
| 176 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.311 |
0.223 |
| 177 |
264489_at |
AT1G27370
|
SPL10, squamosa promoter binding protein-like 10 |
0.310 |
0.003 |
| 178 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.309 |
0.095 |
| 179 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.309 |
0.147 |
| 180 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.308 |
0.700 |
| 181 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.308 |
0.270 |
| 182 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.306 |
-0.129 |
| 183 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.305 |
0.095 |
| 184 |
262291_at |
AT1G70790
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.300 |
0.071 |
| 185 |
254715_at |
AT4G13550
|
[triglyceride lipases] |
0.299 |
-0.078 |
| 186 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.299 |
0.013 |
| 187 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.298 |
0.232 |
| 188 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.297 |
0.144 |
| 189 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.297 |
0.348 |
| 190 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.297 |
0.042 |
| 191 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.297 |
0.021 |
| 192 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.296 |
0.355 |
| 193 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.295 |
0.240 |
| 194 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.295 |
0.112 |
| 195 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
0.295 |
0.163 |
| 196 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.294 |
0.106 |
| 197 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
0.291 |
-0.084 |
| 198 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.291 |
-0.144 |
| 199 |
245370_at |
AT4G16840
|
unknown |
0.288 |
0.190 |
| 200 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.286 |
-0.236 |
| 201 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.286 |
0.091 |
| 202 |
248487_at |
AT5G51070
|
CLPD, ERD1, EARLY RESPONSIVE TO DEHYDRATION 1, SAG15, SENESCENCE ASSOCIATED GENE 15 |
0.285 |
0.119 |
| 203 |
261447_at |
AT1G21160
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
0.284 |
0.110 |
| 204 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.284 |
-0.013 |
| 205 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.283 |
0.018 |
| 206 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
0.282 |
-0.045 |
| 207 |
259226_at |
AT3G07700
|
[Protein kinase superfamily protein] |
0.282 |
0.069 |
| 208 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.281 |
0.044 |
| 209 |
245353_at |
AT4G16000
|
unknown |
0.281 |
-0.038 |
| 210 |
258939_at |
AT3G10020
|
unknown |
0.280 |
0.239 |
| 211 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.278 |
0.201 |
| 212 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.278 |
-0.242 |
| 213 |
246951_at |
AT5G04880
|
[expressed protein] |
0.278 |
0.209 |
| 214 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.277 |
0.133 |
| 215 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
0.277 |
-0.100 |
| 216 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
0.276 |
0.127 |
| 217 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.276 |
-0.029 |
| 218 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.275 |
0.127 |
| 219 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.275 |
-0.067 |
| 220 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
0.275 |
0.077 |
| 221 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.275 |
0.051 |
| 222 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.274 |
0.039 |
| 223 |
249091_at |
AT5G43860
|
ATCLH2, ARABIDOPSIS THALIANA CHLOROPHYLLASE 2, CLH2, chlorophyllase 2 |
0.274 |
0.316 |
| 224 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.274 |
-0.078 |
| 225 |
257827_at |
AT3G26630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.273 |
0.061 |
| 226 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.273 |
0.540 |
| 227 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.273 |
0.197 |
| 228 |
257860_at |
AT3G13062
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.272 |
0.060 |
| 229 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.272 |
0.287 |
| 230 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
0.271 |
0.086 |
| 231 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
0.271 |
0.110 |
| 232 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.269 |
-0.187 |
| 233 |
264423_at |
AT1G61690
|
[phosphoinositide binding] |
0.268 |
0.086 |
| 234 |
262860_at |
AT1G64810
|
APO1, ACCUMULATION OF PHOTOSYSTEM ONE 1 |
0.268 |
-0.022 |
| 235 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.267 |
0.121 |
| 236 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.266 |
0.157 |
| 237 |
249824_at |
AT5G23380
|
unknown |
0.266 |
0.038 |
| 238 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.266 |
0.294 |
| 239 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.266 |
0.032 |
| 240 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.266 |
0.909 |
| 241 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
0.265 |
0.353 |
| 242 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.265 |
-0.065 |
| 243 |
264908_at |
AT2G17440
|
PIRL5, plant intracellular ras group-related LRR 5 |
0.264 |
0.087 |
| 244 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.264 |
0.131 |
| 245 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
0.262 |
-0.001 |
| 246 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
0.262 |
0.078 |
| 247 |
267178_at |
AT2G37750
|
unknown |
0.261 |
0.142 |
| 248 |
250217_at |
AT5G14120
|
[Major facilitator superfamily protein] |
0.261 |
-0.075 |
| 249 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.261 |
0.211 |
| 250 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.260 |
-0.329 |
| 251 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.259 |
0.193 |
| 252 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.258 |
-0.016 |
| 253 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.258 |
-0.015 |
| 254 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.257 |
-0.053 |
| 255 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
0.257 |
-0.018 |
| 256 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
0.257 |
-0.018 |
| 257 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.256 |
-0.106 |
| 258 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.256 |
0.251 |
| 259 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
0.256 |
-0.143 |
| 260 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
0.254 |
-0.021 |
| 261 |
251403_at |
AT3G60300
|
[RWD domain-containing protein] |
0.254 |
0.129 |
| 262 |
260500_at |
AT2G41705
|
[camphor resistance CrcB family protein] |
0.253 |
0.108 |
| 263 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.253 |
-0.022 |
| 264 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.253 |
-0.023 |
| 265 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.253 |
0.200 |
| 266 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.253 |
-0.023 |
| 267 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.252 |
-0.083 |
| 268 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
0.252 |
-0.035 |
| 269 |
256097_at |
AT1G13670
|
unknown |
0.252 |
-0.001 |
| 270 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.251 |
0.234 |
| 271 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.251 |
0.057 |
| 272 |
263674_at |
AT2G04790
|
unknown |
0.251 |
-0.081 |
| 273 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
0.250 |
-0.030 |
| 274 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.250 |
0.020 |
| 275 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.250 |
0.145 |
| 276 |
253647_at |
AT4G29950
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.249 |
-0.036 |
| 277 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.249 |
0.051 |
| 278 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
0.249 |
0.049 |
| 279 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.248 |
0.025 |
| 280 |
261250_at |
AT1G05890
|
ARI5, ARIADNE 5, ATARI5, ARABIDOPSIS ARIADNE 5 |
0.248 |
0.152 |
| 281 |
253343_at |
AT4G33540
|
[metallo-beta-lactamase family protein] |
0.247 |
-0.069 |
| 282 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
0.247 |
0.026 |
| 283 |
253830_at |
AT4G27652
|
unknown |
0.247 |
0.081 |
| 284 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-1.061 |
-0.070 |
| 285 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.914 |
-0.041 |
| 286 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
-0.890 |
0.057 |
| 287 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.872 |
0.342 |
| 288 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.791 |
-0.239 |
| 289 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.765 |
0.096 |
| 290 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.759 |
-0.281 |
| 291 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.748 |
-0.121 |
| 292 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.746 |
-0.542 |
| 293 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.708 |
-0.076 |
| 294 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.699 |
-0.008 |
| 295 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.682 |
-0.005 |
| 296 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.677 |
0.283 |
| 297 |
266222_at |
AT2G28780
|
unknown |
-0.662 |
-0.061 |
| 298 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.652 |
-0.029 |
| 299 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.642 |
-0.091 |
| 300 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.631 |
-0.076 |
| 301 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.630 |
-0.038 |
| 302 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.629 |
-0.031 |
| 303 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.610 |
-0.123 |
| 304 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.603 |
0.437 |
| 305 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.595 |
-0.242 |
| 306 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.580 |
-0.063 |
| 307 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.573 |
0.152 |
| 308 |
261221_at |
AT1G19960
|
unknown |
-0.570 |
0.030 |
| 309 |
263829_at |
AT2G40435
|
unknown |
-0.567 |
-0.341 |
| 310 |
265083_at |
AT1G03820
|
unknown |
-0.563 |
-0.373 |
| 311 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.548 |
-0.136 |
| 312 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.544 |
-0.176 |
| 313 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.542 |
0.049 |
| 314 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.541 |
0.196 |
| 315 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.539 |
-0.106 |
| 316 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.531 |
-0.133 |
| 317 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.530 |
-0.016 |
| 318 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.524 |
0.014 |
| 319 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.521 |
-0.075 |
| 320 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.521 |
-0.386 |
| 321 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.520 |
-0.025 |
| 322 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.509 |
-0.344 |
| 323 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.502 |
-0.015 |
| 324 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.502 |
0.020 |
| 325 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.498 |
-0.172 |
| 326 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.490 |
0.151 |
| 327 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.487 |
0.070 |
| 328 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.478 |
-0.587 |
| 329 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.477 |
0.014 |
| 330 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.468 |
-0.043 |
| 331 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.468 |
0.405 |
| 332 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.466 |
-0.045 |
| 333 |
259735_at |
AT1G64405
|
unknown |
-0.461 |
0.071 |
| 334 |
256924_at |
AT3G29590
|
AT5MAT |
-0.458 |
0.361 |
| 335 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.446 |
0.686 |
| 336 |
250778_at |
AT5G05500
|
MOP10 |
-0.445 |
-0.041 |
| 337 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.445 |
-0.038 |
| 338 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.443 |
-0.355 |
| 339 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.442 |
-0.014 |
| 340 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.438 |
0.011 |
| 341 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.436 |
-0.139 |
| 342 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.433 |
0.410 |
| 343 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.433 |
-0.050 |
| 344 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.431 |
-0.340 |
| 345 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.425 |
-0.176 |
| 346 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.420 |
0.026 |
| 347 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.420 |
-0.075 |
| 348 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.416 |
0.087 |
| 349 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.416 |
-0.052 |
| 350 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.411 |
-0.148 |
| 351 |
261921_at |
AT1G65900
|
unknown |
-0.411 |
-0.168 |
| 352 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.409 |
0.036 |
| 353 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.409 |
0.320 |
| 354 |
254092_at |
AT4G25090
|
[Riboflavin synthase-like superfamily protein] |
-0.406 |
-0.023 |
| 355 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.401 |
-0.042 |
| 356 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.400 |
-0.155 |
| 357 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.399 |
0.297 |
| 358 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.399 |
-0.062 |
| 359 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.392 |
0.384 |
| 360 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.391 |
-0.132 |
| 361 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.387 |
0.085 |
| 362 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.383 |
-0.242 |
| 363 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.383 |
-0.098 |
| 364 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.383 |
-0.133 |
| 365 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.383 |
-0.101 |
| 366 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.379 |
-0.023 |
| 367 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.378 |
-0.034 |
| 368 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.376 |
-0.035 |
| 369 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.376 |
-0.265 |
| 370 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.372 |
-0.030 |
| 371 |
255957_at |
AT1G22160
|
unknown |
-0.372 |
-0.283 |
| 372 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
-0.372 |
0.084 |
| 373 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.372 |
0.008 |
| 374 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.372 |
-0.062 |
| 375 |
249167_at |
AT5G42860
|
unknown |
-0.370 |
0.000 |
| 376 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.369 |
0.021 |
| 377 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.367 |
0.071 |
| 378 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.364 |
0.015 |
| 379 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
-0.364 |
-0.001 |
| 380 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.362 |
0.052 |
| 381 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.358 |
-0.091 |
| 382 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.356 |
-0.166 |
| 383 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.354 |
-0.167 |
| 384 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.351 |
-0.356 |
| 385 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.351 |
-0.356 |
| 386 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.350 |
-0.047 |
| 387 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
-0.349 |
-0.063 |
| 388 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.349 |
-0.205 |
| 389 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.349 |
-0.091 |
| 390 |
265545_at |
AT2G28250
|
NCRK |
-0.348 |
-0.091 |
| 391 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.347 |
-0.433 |
| 392 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.345 |
-0.108 |
| 393 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.344 |
-0.065 |
| 394 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.342 |
-0.310 |
| 395 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.342 |
-0.488 |
| 396 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.341 |
-0.288 |
| 397 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.339 |
-0.173 |
| 398 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.333 |
0.106 |
| 399 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.333 |
-0.368 |
| 400 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.332 |
0.075 |
| 401 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.328 |
-0.040 |
| 402 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.328 |
0.038 |
| 403 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.327 |
0.013 |
| 404 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.325 |
-0.132 |
| 405 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
-0.323 |
-0.093 |
| 406 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.323 |
-0.013 |
| 407 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.323 |
-0.322 |
| 408 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.323 |
0.140 |
| 409 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.322 |
-0.226 |
| 410 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.321 |
-0.167 |
| 411 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.321 |
-0.076 |
| 412 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.321 |
-0.050 |
| 413 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.319 |
-0.370 |
| 414 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.319 |
0.401 |
| 415 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.317 |
0.570 |
| 416 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.316 |
-0.093 |
| 417 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.316 |
0.026 |
| 418 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.315 |
0.435 |
| 419 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.314 |
-0.182 |
| 420 |
247024_at |
AT5G66985
|
unknown |
-0.314 |
0.144 |
| 421 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.313 |
-0.092 |
| 422 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.312 |
-0.116 |
| 423 |
260582_at |
AT2G47200
|
unknown |
-0.309 |
-0.019 |
| 424 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.309 |
-0.168 |
| 425 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.305 |
-0.214 |
| 426 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.304 |
-0.223 |
| 427 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
-0.303 |
-0.288 |
| 428 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.300 |
-0.078 |
| 429 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.300 |
-0.406 |
| 430 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.300 |
-0.017 |
| 431 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.300 |
-0.173 |
| 432 |
251067_at |
AT5G01910
|
unknown |
-0.299 |
-0.176 |
| 433 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.296 |
0.027 |
| 434 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.295 |
-0.073 |
| 435 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
-0.295 |
0.060 |
| 436 |
267012_at |
AT2G39220
|
PLA IIB, PLP6, PATATIN-like protein 6 |
-0.294 |
-0.036 |
| 437 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.293 |
-0.193 |
| 438 |
249303_at |
AT5G41460
|
unknown |
-0.293 |
-0.073 |
| 439 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.292 |
0.027 |
| 440 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.291 |
-0.028 |
| 441 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.291 |
0.123 |
| 442 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.290 |
-0.062 |
| 443 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.290 |
-0.163 |
| 444 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.289 |
0.190 |
| 445 |
251753_at |
AT3G55760
|
unknown |
-0.286 |
-0.056 |
| 446 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.286 |
-0.085 |
| 447 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.286 |
-0.161 |
| 448 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.286 |
0.110 |
| 449 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.285 |
0.185 |
| 450 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.282 |
-0.070 |
| 451 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.282 |
-0.108 |
| 452 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.281 |
-0.294 |
| 453 |
255876_at |
AT2G40480
|
unknown |
-0.280 |
-0.155 |
| 454 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.279 |
-0.104 |
| 455 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.279 |
-0.034 |
| 456 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.278 |
0.137 |
| 457 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.278 |
0.106 |
| 458 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.278 |
-0.091 |
| 459 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.277 |
-0.137 |
| 460 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
-0.276 |
-0.067 |
| 461 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-0.274 |
-0.111 |
| 462 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.274 |
-0.117 |
| 463 |
267636_at |
AT2G42110
|
unknown |
-0.273 |
-0.224 |
| 464 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.272 |
-0.019 |
| 465 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.272 |
-0.327 |
| 466 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.271 |
0.159 |
| 467 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.269 |
0.049 |
| 468 |
247541_at |
AT5G61660
|
[glycine-rich protein] |
-0.269 |
-0.202 |
| 469 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.269 |
-0.252 |
| 470 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.269 |
-0.181 |
| 471 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.268 |
-0.144 |
| 472 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.268 |
-0.048 |
| 473 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.268 |
-0.159 |
| 474 |
252077_at |
AT3G51720
|
unknown |
-0.266 |
-0.174 |
| 475 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.263 |
-0.110 |
| 476 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.263 |
-0.176 |
| 477 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.262 |
-0.006 |
| 478 |
250438_at |
AT5G10580
|
unknown |
-0.262 |
0.018 |
| 479 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.261 |
-0.016 |
| 480 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.260 |
-0.052 |
| 481 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
-0.259 |
-0.215 |
| 482 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.259 |
-0.150 |
| 483 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.259 |
-0.091 |
| 484 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.258 |
-0.201 |
| 485 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.256 |
-0.061 |
| 486 |
253238_at |
AT4G34480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.256 |
-0.100 |
| 487 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.256 |
-0.204 |
| 488 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.256 |
-0.042 |
| 489 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.256 |
0.134 |
| 490 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.255 |
0.040 |
| 491 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
-0.255 |
-0.354 |
| 492 |
251394_at |
AT3G60900
|
FLA10, FASCICLIN-like arabinogalactan-protein 10 |
-0.254 |
-0.243 |
| 493 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.254 |
-0.096 |
| 494 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.253 |
-0.006 |
| 495 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.251 |
-0.003 |
| 496 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.251 |
-0.037 |
| 497 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.251 |
-0.006 |
| 498 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.250 |
0.072 |
| 499 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.250 |
0.180 |
| 500 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.249 |
-0.104 |
| 501 |
264287_at |
AT1G61930
|
unknown |
-0.249 |
0.192 |
| 502 |
258702_at |
AT3G09730
|
unknown |
-0.249 |
-0.036 |
| 503 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.248 |
-0.148 |
| 504 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.247 |
-0.153 |
| 505 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.247 |
-0.075 |
| 506 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.247 |
-0.097 |
| 507 |
252089_at |
AT3G52110
|
unknown |
-0.247 |
-0.160 |
| 508 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.247 |
-0.088 |
| 509 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.246 |
0.086 |
| 510 |
263544_at |
AT2G21590
|
APL4 |
-0.246 |
-0.175 |
| 511 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.246 |
-0.112 |
| 512 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.244 |
0.132 |
| 513 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.244 |
0.008 |
| 514 |
260332_at |
AT1G70470
|
unknown |
-0.242 |
-0.091 |
| 515 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
-0.241 |
0.068 |
| 516 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.241 |
-0.100 |
| 517 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.241 |
-0.222 |
| 518 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.240 |
0.629 |
| 519 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.239 |
-0.031 |
| 520 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.239 |
-0.123 |
| 521 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
-0.238 |
-0.147 |
| 522 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.237 |
-0.053 |
| 523 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.237 |
-0.103 |
| 524 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.237 |
-0.071 |
| 525 |
256512_at |
AT1G33265
|
[Transmembrane proteins 14C] |
-0.237 |
-0.008 |
| 526 |
248309_at |
AT5G52540
|
unknown |
-0.236 |
-0.173 |
| 527 |
261178_at |
AT1G04760
|
ATVAMP726, vesicle-associated membrane protein 726, VAMP726, vesicle-associated membrane protein 726 |
-0.236 |
-0.169 |
| 528 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.235 |
-0.134 |
| 529 |
253804_at |
AT4G28230
|
unknown |
-0.235 |
-0.167 |
| 530 |
253978_at |
AT4G26660
|
unknown |
-0.235 |
-0.142 |
| 531 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.234 |
-0.197 |
| 532 |
247425_at |
AT5G62550
|
unknown |
-0.234 |
-0.169 |
| 533 |
264987_at |
AT1G27030
|
unknown |
-0.234 |
0.086 |
| 534 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.233 |
-0.064 |
| 535 |
256789_at |
AT3G13672
|
SINA2 |
-0.233 |
-0.036 |
| 536 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
-0.232 |
0.147 |
| 537 |
262558_at |
AT1G31335
|
unknown |
-0.231 |
-0.027 |
| 538 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.230 |
-0.045 |
| 539 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.230 |
0.031 |
| 540 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.229 |
-0.039 |
| 541 |
260329_at |
AT1G80370
|
CYCA2;4, Cyclin A2;4 |
-0.229 |
-0.220 |
| 542 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.228 |
-0.108 |
| 543 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.228 |
-0.064 |
| 544 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.228 |
-0.055 |
| 545 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
-0.227 |
-0.127 |
| 546 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.226 |
-0.146 |
| 547 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.226 |
-0.352 |
| 548 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.226 |
-0.130 |
| 549 |
247667_at |
AT5G60150
|
unknown |
-0.225 |
-0.127 |
| 550 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
-0.225 |
0.050 |
| 551 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.225 |
-0.093 |
| 552 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.224 |
-0.156 |
| 553 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.224 |
-0.032 |
| 554 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
-0.224 |
-0.169 |
| 555 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.222 |
-0.131 |
| 556 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.222 |
0.073 |
| 557 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
-0.222 |
-0.092 |
| 558 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.222 |
-0.101 |
| 559 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.222 |
-0.095 |
| 560 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.221 |
-0.006 |
| 561 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.221 |
-0.111 |
| 562 |
254190_at |
AT4G23885
|
unknown |
-0.221 |
0.053 |
| 563 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.221 |
-0.034 |
| 564 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
-0.220 |
-0.044 |
| 565 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.220 |
-0.098 |