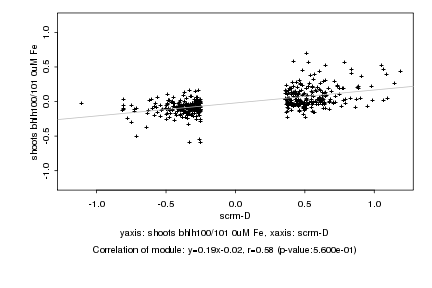
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
scrm-D |
Log2 signal ratio
shoots bhlh100/101 0uM Fe |
| 1 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.186 |
0.438 |
| 2 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
1.144 |
0.268 |
| 3 |
264635_at |
AT1G65500
|
unknown |
1.092 |
0.048 |
| 4 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.084 |
0.399 |
| 5 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
1.065 |
0.024 |
| 6 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.065 |
0.470 |
| 7 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
1.048 |
0.530 |
| 8 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.985 |
0.020 |
| 9 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.975 |
0.222 |
| 10 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.951 |
-0.065 |
| 11 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.904 |
0.376 |
| 12 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.894 |
0.056 |
| 13 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.886 |
0.191 |
| 14 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.881 |
0.230 |
| 15 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.873 |
0.208 |
| 16 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.871 |
0.036 |
| 17 |
267622_at |
AT2G39690
|
unknown |
0.860 |
-0.084 |
| 18 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.835 |
0.475 |
| 19 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.832 |
0.416 |
| 20 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.828 |
0.057 |
| 21 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.792 |
-0.015 |
| 22 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.790 |
0.041 |
| 23 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.784 |
0.577 |
| 24 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.776 |
0.202 |
| 25 |
261146_at |
AT1G19620
|
unknown |
0.772 |
0.015 |
| 26 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.769 |
0.202 |
| 27 |
265002_at |
AT1G24400
|
AATL2, AMINO ACID TRANSPORTER-LIKE PROTEIN 2, ATLHT2, ARABIDOPSIS LYSINE HISTIDINE TRANSPORTER 2, LHT2, lysine histidine transporter 2 |
0.761 |
-0.065 |
| 28 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.746 |
0.107 |
| 29 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.743 |
0.202 |
| 30 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.733 |
0.236 |
| 31 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.728 |
0.219 |
| 32 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.716 |
0.080 |
| 33 |
250614_at |
AT5G07260
|
[START (StAR-related lipid-transfer) lipid-binding domain] |
0.715 |
-0.001 |
| 34 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.713 |
0.293 |
| 35 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
0.706 |
-0.001 |
| 36 |
261012_at |
AT1G26600
|
CLE9, CLAVATA3/ESR-RELATED 9 |
0.702 |
-0.019 |
| 37 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.691 |
0.154 |
| 38 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
0.684 |
0.040 |
| 39 |
263121_at |
AT1G78530
|
[Protein kinase superfamily protein] |
0.677 |
-0.110 |
| 40 |
263465_at |
AT2G31940
|
unknown |
0.672 |
0.001 |
| 41 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.671 |
-0.000 |
| 42 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.669 |
0.197 |
| 43 |
263544_at |
AT2G21590
|
APL4 |
0.652 |
0.099 |
| 44 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.648 |
-0.089 |
| 45 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.648 |
0.525 |
| 46 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.647 |
0.315 |
| 47 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.645 |
0.136 |
| 48 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.645 |
0.084 |
| 49 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.639 |
-0.041 |
| 50 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.639 |
0.117 |
| 51 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.639 |
-0.006 |
| 52 |
266227_at |
AT2G28870
|
unknown |
0.638 |
-0.024 |
| 53 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
0.634 |
-0.092 |
| 54 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.630 |
0.296 |
| 55 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.628 |
0.035 |
| 56 |
263718_at |
AT2G13570
|
NF-YB7, nuclear factor Y, subunit B7 |
0.627 |
0.020 |
| 57 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.626 |
0.137 |
| 58 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.626 |
0.099 |
| 59 |
256395_at |
AT3G06120
|
MUTE, MUTE |
0.625 |
0.055 |
| 60 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.621 |
-0.036 |
| 61 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
0.620 |
-0.034 |
| 62 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
0.617 |
0.013 |
| 63 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
0.616 |
0.033 |
| 64 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.610 |
0.168 |
| 65 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.610 |
0.263 |
| 66 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.609 |
0.021 |
| 67 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.607 |
0.053 |
| 68 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
0.607 |
-0.001 |
| 69 |
266600_at |
AT2G46070
|
ATMPK12, MAPK12, MPK12, mitogen-activated protein kinase 12 |
0.606 |
0.013 |
| 70 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.602 |
0.444 |
| 71 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.601 |
0.072 |
| 72 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.600 |
0.185 |
| 73 |
265587_at |
AT2G19980
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.597 |
-0.034 |
| 74 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.596 |
0.035 |
| 75 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.589 |
0.043 |
| 76 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.587 |
0.207 |
| 77 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.582 |
-0.035 |
| 78 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.581 |
0.151 |
| 79 |
262480_at |
AT1G11340
|
[S-locus lectin protein kinase family protein] |
0.580 |
-0.010 |
| 80 |
266877_at |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
0.572 |
0.011 |
| 81 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.570 |
0.163 |
| 82 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.569 |
0.392 |
| 83 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.563 |
-0.157 |
| 84 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.562 |
0.079 |
| 85 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.560 |
0.325 |
| 86 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.560 |
-0.007 |
| 87 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.556 |
-0.151 |
| 88 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.555 |
0.136 |
| 89 |
247462_at |
AT5G62080
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.555 |
-0.029 |
| 90 |
264958_at |
AT1G76960
|
unknown |
0.554 |
-0.131 |
| 91 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.549 |
0.216 |
| 92 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.548 |
0.227 |
| 93 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.548 |
0.212 |
| 94 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.546 |
-0.003 |
| 95 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.545 |
0.149 |
| 96 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.545 |
-0.007 |
| 97 |
257396_at |
AT2G20875
|
EPF1, EPIDERMAL PATTERNING FACTOR 1 |
0.544 |
-0.074 |
| 98 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
0.544 |
0.041 |
| 99 |
257247_at |
AT3G24140
|
FMA, FAMA |
0.543 |
0.012 |
| 100 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.542 |
0.094 |
| 101 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.535 |
0.385 |
| 102 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.532 |
0.269 |
| 103 |
246297_at |
AT3G51760
|
unknown |
0.531 |
-0.006 |
| 104 |
262531_at |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
0.526 |
-0.035 |
| 105 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.525 |
0.224 |
| 106 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.522 |
0.574 |
| 107 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.515 |
-0.022 |
| 108 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.513 |
0.089 |
| 109 |
263791_at |
AT2G24520
|
AHA5, H(+)-ATPase 5, HA5, H(+)-ATPase 5 |
0.511 |
0.019 |
| 110 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.511 |
0.200 |
| 111 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.510 |
0.699 |
| 112 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.508 |
-0.050 |
| 113 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
0.507 |
-0.091 |
| 114 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.506 |
0.050 |
| 115 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.506 |
0.032 |
| 116 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.505 |
0.199 |
| 117 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.505 |
0.079 |
| 118 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.504 |
-0.048 |
| 119 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.503 |
-0.100 |
| 120 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.498 |
-0.225 |
| 121 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.496 |
0.121 |
| 122 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.496 |
-0.005 |
| 123 |
263830_at |
AT2G40260
|
[Homeodomain-like superfamily protein] |
0.496 |
-0.121 |
| 124 |
266866_at |
AT2G29940
|
ABCG31, ATP-binding cassette G31, ATPDR3, PLEIOTROPIC DRUG RESISTANCE 3, PDR3, pleiotropic drug resistance 3 |
0.494 |
-0.012 |
| 125 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.493 |
-0.005 |
| 126 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
0.489 |
-0.040 |
| 127 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.488 |
-0.061 |
| 128 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.488 |
0.109 |
| 129 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
0.487 |
-0.177 |
| 130 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.486 |
-0.082 |
| 131 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
0.484 |
-0.017 |
| 132 |
264343_at |
AT1G11850
|
unknown |
0.483 |
-0.095 |
| 133 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.479 |
0.235 |
| 134 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.478 |
0.456 |
| 135 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.478 |
0.099 |
| 136 |
254494_at |
AT4G20050
|
QRT3, QUARTET 3 |
0.476 |
-0.026 |
| 137 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.473 |
0.170 |
| 138 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.473 |
0.005 |
| 139 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.467 |
-0.042 |
| 140 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.467 |
0.249 |
| 141 |
261350_at |
AT1G79770
|
unknown |
0.466 |
0.008 |
| 142 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.466 |
0.021 |
| 143 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.462 |
0.264 |
| 144 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.460 |
0.313 |
| 145 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
0.459 |
0.013 |
| 146 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.456 |
-0.136 |
| 147 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.455 |
0.157 |
| 148 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.455 |
-0.121 |
| 149 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.455 |
-0.038 |
| 150 |
266824_at |
AT2G22800
|
HAT9 |
0.452 |
-0.061 |
| 151 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.451 |
0.127 |
| 152 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.451 |
0.114 |
| 153 |
264947_at |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.450 |
0.032 |
| 154 |
257672_at |
AT3G20300
|
unknown |
0.450 |
-0.042 |
| 155 |
265012_at |
AT1G24470
|
ATKCR2, KCR2, beta-ketoacyl reductase 2 |
0.449 |
-0.052 |
| 156 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.447 |
-0.034 |
| 157 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.446 |
0.115 |
| 158 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.445 |
0.078 |
| 159 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.444 |
-0.049 |
| 160 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.444 |
0.216 |
| 161 |
262788_at |
AT1G10690
|
unknown |
0.440 |
0.001 |
| 162 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.438 |
0.013 |
| 163 |
260264_at |
AT1G68500
|
unknown |
0.437 |
0.062 |
| 164 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.434 |
0.125 |
| 165 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.434 |
0.277 |
| 166 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.433 |
-0.025 |
| 167 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.431 |
0.038 |
| 168 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.429 |
0.147 |
| 169 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.428 |
0.103 |
| 170 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
0.428 |
-0.050 |
| 171 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
0.421 |
0.141 |
| 172 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.420 |
-0.042 |
| 173 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.418 |
0.593 |
| 174 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.418 |
-0.058 |
| 175 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.414 |
0.073 |
| 176 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.413 |
0.133 |
| 177 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.412 |
-0.031 |
| 178 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.411 |
0.225 |
| 179 |
247800_at |
AT5G58570
|
unknown |
0.410 |
0.278 |
| 180 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.410 |
-0.039 |
| 181 |
247802_at |
AT5G58580
|
ATL63, TOXICOS EN LEVADURA 63, TL63, TOXICOS EN LEVADURA 63 |
0.409 |
-0.055 |
| 182 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.408 |
0.124 |
| 183 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
0.403 |
-0.020 |
| 184 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.402 |
0.194 |
| 185 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.401 |
0.106 |
| 186 |
247020_at |
AT5G67020
|
unknown |
0.401 |
-0.017 |
| 187 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
0.400 |
-0.051 |
| 188 |
259520_at |
AT1G12320
|
unknown |
0.400 |
0.016 |
| 189 |
256599_at |
AT3G14760
|
unknown |
0.398 |
-0.117 |
| 190 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.397 |
0.134 |
| 191 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.397 |
0.131 |
| 192 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.397 |
0.134 |
| 193 |
249851_at |
AT5G23260
|
ABS, ARABIDOPSIS BSISTER, AGL32, AGAMOUS-like 32, TT16, TRANSPARENT TESTA16 |
0.397 |
-0.033 |
| 194 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.397 |
-0.012 |
| 195 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.393 |
0.190 |
| 196 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.390 |
0.016 |
| 197 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.388 |
-0.023 |
| 198 |
253934_at |
AT4G26830
|
[O-Glycosyl hydrolases family 17 protein] |
0.387 |
-0.042 |
| 199 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.385 |
0.050 |
| 200 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.384 |
0.189 |
| 201 |
255467_at |
AT4G03010
|
[RNI-like superfamily protein] |
0.382 |
-0.055 |
| 202 |
248627_at |
AT5G48950
|
DHNAT2, DHNA-CoA thioesterase 2 |
0.382 |
0.045 |
| 203 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
0.381 |
-0.087 |
| 204 |
265277_at |
AT2G28410
|
unknown |
0.379 |
-0.038 |
| 205 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.379 |
0.124 |
| 206 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.379 |
0.165 |
| 207 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.375 |
0.160 |
| 208 |
256900_at |
AT3G24670
|
[Pectin lyase-like superfamily protein] |
0.375 |
-0.032 |
| 209 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.375 |
-0.003 |
| 210 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.373 |
-0.131 |
| 211 |
246860_at |
AT5G25840
|
unknown |
0.373 |
0.045 |
| 212 |
264012_at |
AT2G21080
|
unknown |
0.373 |
-0.007 |
| 213 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
0.373 |
-0.026 |
| 214 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.371 |
-0.028 |
| 215 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.370 |
-0.081 |
| 216 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.370 |
0.014 |
| 217 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.369 |
0.112 |
| 218 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.369 |
-0.223 |
| 219 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.367 |
0.042 |
| 220 |
259514_at |
AT1G12480
|
CDI3, CARBON DIOXIDE INSENSITIVE 3, OZS1, OZONE-SENSITIVE 1, RCD3, RADICAL-INDUCED CELL DEATH 3, SLAC1, SLOW ANION CHANNEL-ASSOCIATED 1 |
0.364 |
-0.151 |
| 221 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
0.363 |
-0.038 |
| 222 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.362 |
0.232 |
| 223 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.362 |
0.158 |
| 224 |
251535_at |
AT3G58540
|
unknown |
0.360 |
0.034 |
| 225 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.359 |
0.118 |
| 226 |
253168_at |
AT4G35070
|
[SBP (S-ribonuclease binding protein) family protein] |
0.358 |
0.034 |
| 227 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.357 |
0.160 |
| 228 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.354 |
0.057 |
| 229 |
267459_at |
AT2G33850
|
unknown |
-1.113 |
-0.025 |
| 230 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.816 |
-0.119 |
| 231 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.813 |
0.032 |
| 232 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.813 |
-0.115 |
| 233 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.811 |
-0.054 |
| 234 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.809 |
-0.093 |
| 235 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.778 |
-0.241 |
| 236 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.752 |
-0.056 |
| 237 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.752 |
-0.301 |
| 238 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.732 |
-0.105 |
| 239 |
266222_at |
AT2G28780
|
unknown |
-0.729 |
-0.138 |
| 240 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.718 |
-0.091 |
| 241 |
256096_at |
AT1G13650
|
unknown |
-0.714 |
-0.498 |
| 242 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.647 |
-0.363 |
| 243 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.634 |
-0.161 |
| 244 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.632 |
-0.128 |
| 245 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.625 |
0.018 |
| 246 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.611 |
0.038 |
| 247 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.599 |
-0.099 |
| 248 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.590 |
-0.191 |
| 249 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.587 |
-0.061 |
| 250 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.577 |
-0.023 |
| 251 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.573 |
-0.073 |
| 252 |
251058_at |
AT5G01790
|
unknown |
-0.568 |
-0.151 |
| 253 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.568 |
0.072 |
| 254 |
250936_at |
AT5G03120
|
unknown |
-0.541 |
-0.050 |
| 255 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.541 |
-0.213 |
| 256 |
267344_at |
AT2G44230
|
unknown |
-0.536 |
-0.117 |
| 257 |
265892_at |
AT2G15020
|
unknown |
-0.530 |
-0.112 |
| 258 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-0.508 |
-0.100 |
| 259 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.503 |
-0.176 |
| 260 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.501 |
-0.112 |
| 261 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.500 |
-0.247 |
| 262 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.497 |
-0.050 |
| 263 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.494 |
-0.169 |
| 264 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.493 |
0.109 |
| 265 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.492 |
-0.025 |
| 266 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.490 |
-0.035 |
| 267 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.487 |
-0.092 |
| 268 |
256924_at |
AT3G29590
|
AT5MAT |
-0.483 |
0.028 |
| 269 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.483 |
0.021 |
| 270 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.479 |
-0.223 |
| 271 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.478 |
-0.120 |
| 272 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.475 |
0.017 |
| 273 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.474 |
-0.163 |
| 274 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.473 |
-0.104 |
| 275 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.466 |
-0.008 |
| 276 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.459 |
0.004 |
| 277 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.459 |
-0.124 |
| 278 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.458 |
-0.108 |
| 279 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.451 |
-0.062 |
| 280 |
255028_at |
AT4G09890
|
unknown |
-0.450 |
-0.015 |
| 281 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.450 |
0.064 |
| 282 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.449 |
-0.061 |
| 283 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.444 |
-0.065 |
| 284 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.443 |
-0.262 |
| 285 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.441 |
-0.028 |
| 286 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.435 |
-0.173 |
| 287 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.429 |
-0.085 |
| 288 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.426 |
-0.201 |
| 289 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.422 |
-0.142 |
| 290 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.420 |
-0.076 |
| 291 |
258196_at |
AT3G13980
|
unknown |
-0.415 |
-0.102 |
| 292 |
247474_at |
AT5G62280
|
unknown |
-0.412 |
-0.016 |
| 293 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.411 |
-0.063 |
| 294 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.405 |
-0.080 |
| 295 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.404 |
-0.011 |
| 296 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.403 |
-0.061 |
| 297 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
-0.403 |
-0.042 |
| 298 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.401 |
-0.197 |
| 299 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.400 |
0.024 |
| 300 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.399 |
-0.073 |
| 301 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.398 |
-0.120 |
| 302 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.396 |
-0.186 |
| 303 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.396 |
-0.104 |
| 304 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.395 |
-0.080 |
| 305 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.391 |
-0.047 |
| 306 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.391 |
-0.110 |
| 307 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.388 |
-0.103 |
| 308 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.387 |
-0.072 |
| 309 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.385 |
-0.229 |
| 310 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.381 |
-0.092 |
| 311 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.379 |
-0.162 |
| 312 |
252215_at |
AT3G50240
|
KICP-02 |
-0.377 |
-0.130 |
| 313 |
265543_at |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.376 |
-0.048 |
| 314 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.376 |
-0.142 |
| 315 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.376 |
0.093 |
| 316 |
247878_at |
AT5G57760
|
unknown |
-0.376 |
-0.137 |
| 317 |
248942_at |
AT5G45480
|
unknown |
-0.375 |
-0.035 |
| 318 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.371 |
-0.142 |
| 319 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.370 |
0.135 |
| 320 |
263002_at |
AT1G54200
|
unknown |
-0.368 |
0.001 |
| 321 |
262598_at |
AT1G15260
|
unknown |
-0.366 |
-0.034 |
| 322 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.363 |
-0.217 |
| 323 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.361 |
-0.130 |
| 324 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.358 |
-0.169 |
| 325 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.358 |
-0.023 |
| 326 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.357 |
-0.058 |
| 327 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.354 |
0.044 |
| 328 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.354 |
-0.031 |
| 329 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.353 |
-0.145 |
| 330 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.353 |
-0.090 |
| 331 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.352 |
-0.234 |
| 332 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.350 |
-0.084 |
| 333 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.348 |
-0.148 |
| 334 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.348 |
-0.241 |
| 335 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.348 |
-0.197 |
| 336 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.346 |
-0.071 |
| 337 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.345 |
-0.103 |
| 338 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.344 |
-0.037 |
| 339 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.343 |
-0.145 |
| 340 |
255957_at |
AT1G22160
|
unknown |
-0.342 |
-0.323 |
| 341 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.337 |
0.164 |
| 342 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.337 |
-0.584 |
| 343 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.336 |
-0.140 |
| 344 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.333 |
-0.049 |
| 345 |
264909_at |
AT2G17300
|
unknown |
-0.333 |
-0.136 |
| 346 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.331 |
-0.178 |
| 347 |
256761_at |
AT3G25670
|
[Leucine-rich repeat (LRR) family protein] |
-0.330 |
-0.069 |
| 348 |
251284_at |
AT3G61840
|
unknown |
-0.328 |
-0.036 |
| 349 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.328 |
-0.069 |
| 350 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.327 |
-0.081 |
| 351 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.327 |
-0.042 |
| 352 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.324 |
0.081 |
| 353 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.324 |
-0.114 |
| 354 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.324 |
-0.096 |
| 355 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.323 |
-0.059 |
| 356 |
259001_at |
AT3G01960
|
unknown |
-0.323 |
-0.195 |
| 357 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.322 |
-0.060 |
| 358 |
260696_at |
AT1G32520
|
unknown |
-0.321 |
-0.100 |
| 359 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.320 |
-0.090 |
| 360 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.319 |
-0.115 |
| 361 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.318 |
-0.104 |
| 362 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.318 |
0.075 |
| 363 |
257221_at |
AT3G27920
|
ATGL1, ATMYB0, myb domain protein 0, GL1, GLABRA 1, MYB0, myb domain protein 0 |
-0.318 |
-0.129 |
| 364 |
252054_at |
AT3G52540
|
ATOFP18, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 18, OFP18, ovate family protein 18 |
-0.317 |
-0.191 |
| 365 |
253800_at |
AT4G28160
|
[hydroxyproline-rich glycoprotein family protein] |
-0.316 |
-0.078 |
| 366 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.315 |
-0.028 |
| 367 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.315 |
-0.055 |
| 368 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.315 |
-0.102 |
| 369 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.311 |
-0.089 |
| 370 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.311 |
-0.161 |
| 371 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.311 |
-0.015 |
| 372 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.310 |
-0.023 |
| 373 |
260363_at |
AT1G70550
|
unknown |
-0.308 |
-0.119 |
| 374 |
258017_at |
AT3G19370
|
unknown |
-0.308 |
-0.042 |
| 375 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.308 |
-0.063 |
| 376 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.306 |
-0.126 |
| 377 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.305 |
-0.205 |
| 378 |
262446_at |
AT1G49310
|
unknown |
-0.304 |
-0.049 |
| 379 |
261536_at |
AT1G01790
|
ATKEA1, K+ EFFLUX ANTIPORTER 1, KEA1, K+ efflux antiporter 1 |
-0.303 |
-0.073 |
| 380 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.303 |
-0.079 |
| 381 |
252050_at |
AT3G52550
|
unknown |
-0.303 |
-0.103 |
| 382 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.302 |
-0.016 |
| 383 |
250734_at |
AT5G06270
|
unknown |
-0.302 |
-0.132 |
| 384 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.301 |
-0.126 |
| 385 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.301 |
-0.112 |
| 386 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.301 |
-0.109 |
| 387 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.299 |
-0.163 |
| 388 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.298 |
-0.131 |
| 389 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.298 |
0.037 |
| 390 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.298 |
-0.117 |
| 391 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.295 |
-0.071 |
| 392 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.294 |
0.010 |
| 393 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.294 |
-0.084 |
| 394 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.292 |
-0.095 |
| 395 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.291 |
-0.022 |
| 396 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.288 |
0.155 |
| 397 |
260012_at |
AT1G67865
|
unknown |
-0.287 |
-0.001 |
| 398 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.287 |
-0.017 |
| 399 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.285 |
0.005 |
| 400 |
246231_at |
AT4G37080
|
unknown |
-0.285 |
-0.087 |
| 401 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.285 |
-0.089 |
| 402 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
-0.284 |
-0.107 |
| 403 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.284 |
-0.121 |
| 404 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.283 |
-0.258 |
| 405 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.282 |
-0.080 |
| 406 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.281 |
0.063 |
| 407 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.281 |
-0.214 |
| 408 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.281 |
-0.089 |
| 409 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.281 |
-0.037 |
| 410 |
250284_at |
AT5G13290
|
CRN, CORYNE, SOL2, SUPPRESSOR OF LLP1 2 |
-0.280 |
-0.035 |
| 411 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.280 |
-0.125 |
| 412 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.280 |
0.059 |
| 413 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.279 |
-0.045 |
| 414 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.278 |
-0.044 |
| 415 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.275 |
0.042 |
| 416 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.274 |
-0.000 |
| 417 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.274 |
-0.093 |
| 418 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
-0.274 |
-0.067 |
| 419 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.273 |
-0.045 |
| 420 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.273 |
-0.034 |
| 421 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.272 |
-0.160 |
| 422 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
-0.272 |
-0.128 |
| 423 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.271 |
-0.121 |
| 424 |
247024_at |
AT5G66985
|
unknown |
-0.271 |
0.160 |
| 425 |
256799_at |
AT3G18560
|
unknown |
-0.270 |
-0.009 |
| 426 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.269 |
-0.052 |
| 427 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.269 |
-0.134 |
| 428 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
-0.269 |
-0.067 |
| 429 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.268 |
-0.151 |
| 430 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.268 |
-0.067 |
| 431 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.267 |
-0.148 |
| 432 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.266 |
-0.545 |
| 433 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.266 |
-0.051 |
| 434 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.265 |
-0.129 |
| 435 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.265 |
-0.121 |
| 436 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.264 |
-0.028 |
| 437 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.264 |
-0.120 |
| 438 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.263 |
-0.151 |
| 439 |
256066_at |
AT1G06980
|
unknown |
-0.260 |
-0.188 |
| 440 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.259 |
0.018 |
| 441 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.257 |
-0.117 |
| 442 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.257 |
-0.585 |
| 443 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
-0.256 |
0.011 |
| 444 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.256 |
0.030 |
| 445 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.256 |
-0.111 |
| 446 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.256 |
-0.247 |
| 447 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.256 |
-0.105 |
| 448 |
263674_at |
AT2G04790
|
unknown |
-0.255 |
-0.020 |
| 449 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.254 |
-0.005 |
| 450 |
245726_at |
AT1G73360
|
AtEDT1, ATHDG11, HOMEODOMAIN GLABROUS 11, EDT1, ENHANCED DROUGHT TOLERANCE 1, HDG11, homeodomain GLABROUS 11 |
-0.253 |
-0.132 |
| 451 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.253 |
-0.286 |
| 452 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.253 |
-0.063 |
| 453 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.253 |
-0.106 |
| 454 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
-0.253 |
-0.018 |
| 455 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.253 |
-0.077 |