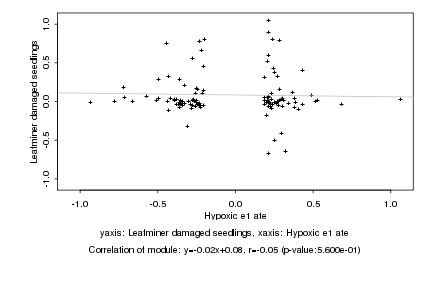
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Hypoxic e1 ate |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
245226_at |
AT3G29970
|
[B12D protein] |
1.060 |
0.029 |
| 2 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.680 |
-0.037 |
| 3 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.522 |
0.016 |
| 4 |
266222_at |
AT2G28780
|
unknown |
0.510 |
0.005 |
| 5 |
252882_at |
AT4G39675
|
unknown |
0.486 |
0.080 |
| 6 |
250472_at |
AT5G10210
|
unknown |
0.427 |
-0.029 |
| 7 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.427 |
0.413 |
| 8 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.402 |
-0.103 |
| 9 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
0.381 |
-0.000 |
| 10 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
0.376 |
0.043 |
| 11 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.375 |
-0.077 |
| 12 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.364 |
0.120 |
| 13 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
0.337 |
-0.017 |
| 14 |
246001_at |
AT5G20790
|
unknown |
0.319 |
-0.646 |
| 15 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.303 |
0.024 |
| 16 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.303 |
0.021 |
| 17 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.302 |
-0.058 |
| 18 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.301 |
0.048 |
| 19 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.296 |
-0.404 |
| 20 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.292 |
0.039 |
| 21 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.282 |
0.017 |
| 22 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.281 |
0.792 |
| 23 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.280 |
0.162 |
| 24 |
267580_at |
AT2G41990
|
unknown |
0.277 |
-0.044 |
| 25 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.272 |
0.008 |
| 26 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.266 |
0.333 |
| 27 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
0.265 |
0.003 |
| 28 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
0.261 |
-0.021 |
| 29 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.258 |
-0.019 |
| 30 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.250 |
-0.009 |
| 31 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.249 |
-0.495 |
| 32 |
258362_at |
AT3G14280
|
unknown |
0.248 |
0.383 |
| 33 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.242 |
0.436 |
| 34 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.236 |
0.813 |
| 35 |
259735_at |
AT1G64405
|
unknown |
0.236 |
-0.028 |
| 36 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.231 |
-0.085 |
| 37 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.227 |
0.029 |
| 38 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.226 |
0.108 |
| 39 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
0.221 |
-0.052 |
| 40 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.215 |
-0.014 |
| 41 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.213 |
-0.010 |
| 42 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.211 |
0.072 |
| 43 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.211 |
0.603 |
| 44 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.210 |
0.894 |
| 45 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.210 |
-0.671 |
| 46 |
260668_at |
AT1G19530
|
unknown |
0.210 |
0.053 |
| 47 |
262446_at |
AT1G49310
|
unknown |
0.209 |
0.001 |
| 48 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.208 |
1.055 |
| 49 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.208 |
-0.062 |
| 50 |
250438_at |
AT5G10580
|
unknown |
0.206 |
0.015 |
| 51 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.205 |
0.524 |
| 52 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.201 |
0.053 |
| 53 |
246911_at |
AT5G25810
|
tny, TINY |
0.197 |
-0.000 |
| 54 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.194 |
-0.168 |
| 55 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.186 |
-0.032 |
| 56 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.186 |
0.063 |
| 57 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.183 |
0.015 |
| 58 |
249136_at |
AT5G43180
|
unknown |
0.182 |
0.317 |
| 59 |
250754_at |
AT5G05700
|
ATATE1, ATE1, arginine-tRNA protein transferase 1, DLS1, DELAYED LEAF SENESCENCE 1 |
-0.939 |
-0.007 |
| 60 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.782 |
0.002 |
| 61 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.724 |
0.189 |
| 62 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.717 |
0.055 |
| 63 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.663 |
0.010 |
| 64 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.578 |
0.075 |
| 65 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
-0.513 |
0.016 |
| 66 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.500 |
0.293 |
| 67 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.497 |
0.048 |
| 68 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.448 |
0.758 |
| 69 |
256257_at |
AT3G11240
|
ATATE2, ATE2, arginine-tRNA protein transferase 2 |
-0.439 |
0.012 |
| 70 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.433 |
0.324 |
| 71 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.431 |
-0.107 |
| 72 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.419 |
0.046 |
| 73 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.397 |
0.020 |
| 74 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.394 |
0.036 |
| 75 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
-0.385 |
-0.029 |
| 76 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.382 |
0.034 |
| 77 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.369 |
-0.032 |
| 78 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.365 |
0.288 |
| 79 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
-0.365 |
-0.069 |
| 80 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.361 |
-0.009 |
| 81 |
259653_at |
AT1G55240
|
unknown |
-0.360 |
0.015 |
| 82 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.359 |
-0.026 |
| 83 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.346 |
-0.039 |
| 84 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.344 |
0.010 |
| 85 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.342 |
-0.025 |
| 86 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.331 |
0.211 |
| 87 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.329 |
-0.023 |
| 88 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.313 |
-0.318 |
| 89 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.303 |
0.012 |
| 90 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.290 |
-0.033 |
| 91 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.287 |
-0.079 |
| 92 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.280 |
-0.043 |
| 93 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.279 |
0.014 |
| 94 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
-0.279 |
0.559 |
| 95 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-0.273 |
0.027 |
| 96 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.267 |
0.005 |
| 97 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
-0.263 |
0.105 |
| 98 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
-0.260 |
-0.058 |
| 99 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.254 |
0.178 |
| 100 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
-0.253 |
-0.006 |
| 101 |
253616_at |
AT4G30380
|
[Barwin-related endoglucanase] |
-0.252 |
0.024 |
| 102 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.249 |
-0.036 |
| 103 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.245 |
0.168 |
| 104 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.241 |
-0.029 |
| 105 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.236 |
0.780 |
| 106 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.233 |
-0.016 |
| 107 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.232 |
-0.040 |
| 108 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.227 |
-0.044 |
| 109 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.225 |
-0.068 |
| 110 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.222 |
0.672 |
| 111 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.215 |
0.107 |
| 112 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.212 |
0.454 |
| 113 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.212 |
0.153 |
| 114 |
247800_at |
AT5G58570
|
unknown |
-0.210 |
-0.044 |
| 115 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.201 |
0.807 |