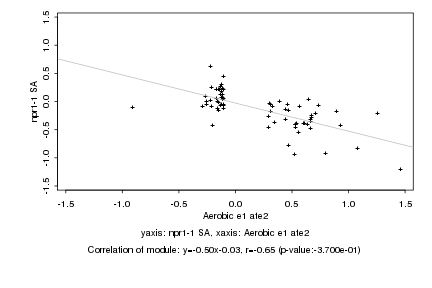
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Aerobic e1 ate2 |
Log2 signal ratio
npr1-1 SA |
| 1 |
245226_at |
AT3G29970
|
[B12D protein] |
1.456 |
-1.197 |
| 2 |
252882_at |
AT4G39675
|
unknown |
1.248 |
-0.196 |
| 3 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.079 |
-0.819 |
| 4 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.926 |
-0.420 |
| 5 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.889 |
-0.175 |
| 6 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.794 |
-0.906 |
| 7 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.727 |
-0.067 |
| 8 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.706 |
-0.204 |
| 9 |
261567_at |
AT1G33055
|
unknown |
0.672 |
-0.243 |
| 10 |
254200_at |
AT4G24110
|
unknown |
0.667 |
-0.266 |
| 11 |
247024_at |
AT5G66985
|
unknown |
0.663 |
-0.343 |
| 12 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.658 |
-0.307 |
| 13 |
250152_at |
AT5G15120
|
unknown |
0.656 |
-0.469 |
| 14 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.645 |
0.048 |
| 15 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.635 |
-0.401 |
| 16 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.610 |
-0.378 |
| 17 |
249384_at |
AT5G39890
|
unknown |
0.593 |
-0.376 |
| 18 |
250472_at |
AT5G10210
|
unknown |
0.558 |
-0.083 |
| 19 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.549 |
-0.548 |
| 20 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.533 |
-0.379 |
| 21 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.530 |
-0.459 |
| 22 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.527 |
-0.403 |
| 23 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.524 |
-0.397 |
| 24 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.519 |
-0.939 |
| 25 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.468 |
-0.774 |
| 26 |
246001_at |
AT5G20790
|
unknown |
0.464 |
-0.143 |
| 27 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.458 |
-0.051 |
| 28 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.440 |
-0.126 |
| 29 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.437 |
-0.313 |
| 30 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
0.383 |
0.009 |
| 31 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.337 |
-0.360 |
| 32 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.319 |
-0.074 |
| 33 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.308 |
-0.038 |
| 34 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.304 |
-0.174 |
| 35 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
0.299 |
-0.030 |
| 36 |
257925_at |
AT3G23170
|
unknown |
0.292 |
-0.257 |
| 37 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
0.287 |
-0.452 |
| 38 |
250754_at |
AT5G05700
|
ATATE1, ATE1, arginine-tRNA protein transferase 1, DLS1, DELAYED LEAF SENESCENCE 1 |
-0.914 |
-0.089 |
| 39 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.293 |
-0.076 |
| 40 |
267459_at |
AT2G33850
|
unknown |
-0.268 |
0.094 |
| 41 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.258 |
-0.043 |
| 42 |
250882_at |
AT5G04000
|
unknown |
-0.257 |
0.016 |
| 43 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.229 |
0.024 |
| 44 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.228 |
0.635 |
| 45 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.219 |
0.256 |
| 46 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.215 |
-0.083 |
| 47 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.209 |
-0.419 |
| 48 |
255808_at |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
-0.176 |
0.055 |
| 49 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.176 |
0.219 |
| 50 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.170 |
0.228 |
| 51 |
257221_at |
AT3G27920
|
ATGL1, ATMYB0, myb domain protein 0, GL1, GLABRA 1, MYB0, myb domain protein 0 |
-0.163 |
-0.114 |
| 52 |
257158_at |
AT3G24360
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.159 |
0.007 |
| 53 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.158 |
-0.142 |
| 54 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.157 |
0.230 |
| 55 |
246888_at |
AT5G26270
|
unknown |
-0.150 |
-0.004 |
| 56 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.145 |
0.217 |
| 57 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
-0.138 |
0.283 |
| 58 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.136 |
-0.063 |
| 59 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.135 |
0.135 |
| 60 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.130 |
-0.042 |
| 61 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.125 |
0.309 |
| 62 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.124 |
0.081 |
| 63 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.124 |
0.187 |
| 64 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.123 |
0.234 |
| 65 |
266560_at |
AT2G23950
|
[Leucine-rich repeat protein kinase family protein] |
-0.122 |
0.083 |
| 66 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
-0.117 |
0.140 |
| 67 |
262890_at |
AT1G14850
|
NUP155, nucleoporin 155 |
-0.117 |
0.041 |
| 68 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
-0.113 |
0.062 |
| 69 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.112 |
0.216 |
| 70 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.110 |
-0.064 |
| 71 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.107 |
-0.109 |
| 72 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.107 |
0.456 |
| 73 |
259762_at |
AT1G77600
|
[ARM repeat superfamily protein] |
-0.106 |
-0.036 |