|
probeID |
AGICode |
Annotation |
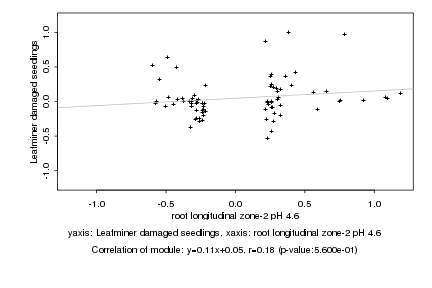
Log2 signal ratio
root longitudinal zone-2 pH 4.6 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
1.186 |
0.127 |
| 2 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
1.094 |
0.053 |
| 3 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.077 |
0.062 |
| 4 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.922 |
0.017 |
| 5 |
264580_at |
AT1G05340
|
unknown |
0.781 |
0.981 |
| 6 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.754 |
0.024 |
| 7 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.749 |
0.003 |
| 8 |
254201_at |
AT4G24130
|
unknown |
0.649 |
0.154 |
| 9 |
258661_at |
AT3G02930
|
unknown |
0.585 |
-0.111 |
| 10 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.556 |
0.132 |
| 11 |
258091_at |
AT3G14560
|
unknown |
0.427 |
0.432 |
| 12 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.403 |
0.243 |
| 13 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.375 |
1.009 |
| 14 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.356 |
0.370 |
| 15 |
257502_at |
AT1G78110
|
unknown |
0.324 |
-0.190 |
| 16 |
250497_at |
AT5G09630
|
[LisH/CRA/RING-U-box domains-containing protein] |
0.324 |
0.176 |
| 17 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.322 |
-0.051 |
| 18 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.303 |
0.060 |
| 19 |
260993_at |
AT1G12140
|
FMO GS-OX5, flavin-monooxygenase glucosinolate S-oxygenase 5 |
0.298 |
0.035 |
| 20 |
249652_at |
AT5G37070
|
unknown |
0.296 |
0.155 |
| 21 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.294 |
0.198 |
| 22 |
247628_at |
AT5G60400
|
unknown |
0.275 |
-0.161 |
| 23 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.271 |
-0.287 |
| 24 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.269 |
0.215 |
| 25 |
245073_at |
AT2G23210
|
[UDP-Glycosyltransferase superfamily protein] |
0.261 |
-0.073 |
| 26 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.259 |
0.399 |
| 27 |
260951_at |
AT1G06150
|
EMB1444, EMBRYO DEFECTIVE 1444 |
0.257 |
-0.072 |
| 28 |
263632_at |
AT2G04795
|
unknown |
0.257 |
-0.423 |
| 29 |
262755_at |
AT1G16360
|
[LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein] |
0.255 |
-0.010 |
| 30 |
263703_at |
AT1G31170
|
ATSRX, SULFIREDOXIN, SRX, sulfiredoxin |
0.255 |
0.004 |
| 31 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
0.253 |
0.257 |
| 32 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.251 |
0.364 |
| 33 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.249 |
0.229 |
| 34 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.233 |
-0.037 |
| 35 |
259528_at |
AT1G12330
|
unknown |
0.230 |
-0.534 |
| 36 |
247229_at |
AT5G65160
|
TPR14, tetratricopeptide repeat 14 |
0.227 |
0.014 |
| 37 |
266480_at |
AT2G31130
|
unknown |
0.224 |
-0.001 |
| 38 |
250327_at |
AT5G12050
|
unknown |
0.218 |
-0.252 |
| 39 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.213 |
0.872 |
| 40 |
267488_at |
AT2G19110
|
ATHMA4, ARABIDOPSIS HEAVY METAL ATPASE 4, HMA4, heavy metal atpase 4 |
0.213 |
-0.113 |
| 41 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.599 |
0.531 |
| 42 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.579 |
-0.028 |
| 43 |
265683_at |
AT2G24400
|
SAUR38, SMALL AUXIN UPREGULATED RNA 38 |
-0.571 |
0.011 |
| 44 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.552 |
0.325 |
| 45 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.504 |
-0.064 |
| 46 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.492 |
0.643 |
| 47 |
252302_at |
AT3G49190
|
[O-acyltransferase (WSD1-like) family protein] |
-0.486 |
0.065 |
| 48 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.449 |
-0.036 |
| 49 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
-0.425 |
0.500 |
| 50 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.420 |
0.040 |
| 51 |
267164_at |
AT2G37700
|
[Fatty acid hydroxylase superfamily] |
-0.384 |
0.051 |
| 52 |
249766_at |
AT5G24070
|
[Peroxidase superfamily protein] |
-0.379 |
0.009 |
| 53 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.338 |
0.007 |
| 54 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.328 |
-0.367 |
| 55 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.321 |
0.005 |
| 56 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.321 |
-0.024 |
| 57 |
247139_at |
AT5G66090
|
unknown |
-0.318 |
-0.064 |
| 58 |
245303_at |
AT4G17010
|
unknown |
-0.310 |
0.058 |
| 59 |
255535_at |
AT4G01790
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.301 |
0.090 |
| 60 |
252446_at |
AT3G47010
|
[Glycosyl hydrolase family protein] |
-0.289 |
-0.253 |
| 61 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.287 |
-0.236 |
| 62 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.286 |
-0.122 |
| 63 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.285 |
0.029 |
| 64 |
255542_at |
AT4G01860
|
[Transducin family protein / WD-40 repeat family protein] |
-0.281 |
-0.025 |
| 65 |
266210_at |
AT2G27530
|
PGY1, PIGGYBACK1 |
-0.278 |
-0.002 |
| 66 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
-0.273 |
0.031 |
| 67 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.265 |
-0.233 |
| 68 |
252852_at |
AT4G39900
|
unknown |
-0.261 |
-0.287 |
| 69 |
261639_at |
AT1G50010
|
TUA2, tubulin alpha-2 chain |
-0.244 |
-0.122 |
| 70 |
253690_at |
AT4G29550
|
unknown |
-0.241 |
-0.016 |
| 71 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.240 |
-0.268 |
| 72 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.239 |
-0.027 |
| 73 |
265545_at |
AT2G28250
|
NCRK |
-0.239 |
-0.147 |
| 74 |
246738_at |
AT5G27740
|
EMB161, EMBRYO DEFECTIVE 161, EMB251, EMBRYO DEFECTIVE 251, EMB2775, EMBRYO DEFECTIVE 2775, RFC3, replication factor C 3 |
-0.235 |
-0.064 |
| 75 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
-0.232 |
-0.189 |
| 76 |
246274_at |
AT4G36620
|
GATA19, GATA transcription factor 19, HANL2, hanaba taranu like 2 |
-0.232 |
-0.101 |
| 77 |
251805_at |
AT3G55530
|
AtSDIR1, SDIR1, SALT- AND DROUGHT-INDUCED RING FINGER1 |
-0.227 |
-0.016 |
| 78 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.221 |
0.238 |
| 79 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.220 |
-0.131 |