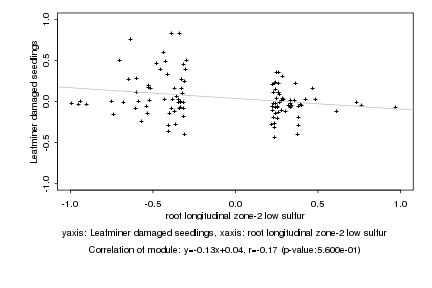
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root longitudinal zone-2 low sulfur |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
247476_at |
AT5G62330
|
unknown |
0.970 |
-0.068 |
| 2 |
250068_at |
AT5G17960
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.764 |
-0.048 |
| 3 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.730 |
-0.004 |
| 4 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.607 |
-0.114 |
| 5 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.481 |
0.033 |
| 6 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.463 |
0.167 |
| 7 |
254213_at |
AT4G23590
|
[Tyrosine transaminase family protein] |
0.420 |
0.033 |
| 8 |
263923_at |
AT2G36485
|
[ENTH/VHS family protein] |
0.395 |
-0.047 |
| 9 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.393 |
-0.027 |
| 10 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
0.381 |
-0.186 |
| 11 |
261540_at |
AT1G63610
|
unknown |
0.379 |
-0.054 |
| 12 |
248423_at |
AT5G51670
|
unknown |
0.377 |
-0.289 |
| 13 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.376 |
-0.396 |
| 14 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.361 |
0.220 |
| 15 |
258638_at |
AT3G07950
|
[rhomboid protein-related] |
0.352 |
0.013 |
| 16 |
253465_at |
AT4G32120
|
[Galactosyltransferase family protein] |
0.335 |
-0.038 |
| 17 |
257613_at |
AT3G26610
|
[Pectin lyase-like superfamily protein] |
0.332 |
-0.014 |
| 18 |
249728_at |
AT5G24390
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.331 |
-0.058 |
| 19 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.331 |
-0.061 |
| 20 |
255318_at |
AT4G04190
|
unknown |
0.329 |
0.023 |
| 21 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.320 |
-0.046 |
| 22 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
0.301 |
-0.121 |
| 23 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.290 |
0.033 |
| 24 |
257670_at |
AT3G20340
|
unknown |
0.283 |
0.311 |
| 25 |
255267_at |
AT4G05210
|
AtLpxD1, LpxD1, lipid X D1 |
0.281 |
0.037 |
| 26 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.278 |
0.015 |
| 27 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
0.274 |
-0.104 |
| 28 |
262222_at |
AT1G74700
|
NUZ, TRZ1, tRNAse Z1 |
0.261 |
-0.007 |
| 29 |
248778_at |
AT5G47940
|
unknown |
0.261 |
0.097 |
| 30 |
250127_at |
AT5G16380
|
unknown |
0.260 |
0.361 |
| 31 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.260 |
0.113 |
| 32 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
0.259 |
0.223 |
| 33 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.256 |
-0.122 |
| 34 |
261852_at |
AT1G50440
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.255 |
0.121 |
| 35 |
245031_at |
AT2G26360
|
[Mitochondrial substrate carrier family protein] |
0.250 |
-0.052 |
| 36 |
250838_at |
AT5G04630
|
CYP77A9, cytochrome P450, family 77, subfamily A, polypeptide 9 |
0.249 |
-0.061 |
| 37 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
0.249 |
-0.206 |
| 38 |
248874_at |
AT5G46460
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.246 |
0.040 |
| 39 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.245 |
0.362 |
| 40 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.241 |
-0.018 |
| 41 |
251840_at |
AT3G54960
|
ATPDI1, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 1, ATPDIL1-3, PDI-like 1-3, PDI1, PROTEIN DISULFIDE ISOMERASE 1, PDIL1-3, PDI-like 1-3 |
0.240 |
0.241 |
| 42 |
260951_at |
AT1G06150
|
EMB1444, EMBRYO DEFECTIVE 1444 |
0.239 |
-0.072 |
| 43 |
252241_at |
AT3G50050
|
[Eukaryotic aspartyl protease family protein] |
0.237 |
-0.134 |
| 44 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.237 |
0.158 |
| 45 |
254815_at |
AT4G12420
|
SKU5 |
0.235 |
-0.432 |
| 46 |
266334_at |
AT2G32380
|
[Transmembrane protein 97, predicted] |
0.233 |
0.222 |
| 47 |
249060_at |
AT5G44560
|
VPS2.2 |
0.233 |
-0.311 |
| 48 |
264779_at |
AT1G08570
|
ACHT4, atypical CYS HIS rich thioredoxin 4 |
0.233 |
-0.260 |
| 49 |
265457_at |
AT2G46550
|
unknown |
0.229 |
0.113 |
| 50 |
265650_at |
AT2G27460
|
[sec23/sec24 transport family protein] |
0.228 |
-0.014 |
| 51 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
0.226 |
-0.194 |
| 52 |
249334_at |
AT5G41000
|
AtYSL4, YSL4, YELLOW STRIPE like 4 |
0.223 |
-0.098 |
| 53 |
262647_at |
AT1G14020
|
[O-fucosyltransferase family protein] |
0.223 |
-0.051 |
| 54 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
0.220 |
0.218 |
| 55 |
255055_at |
AT4G09810
|
[Nucleotide-sugar transporter family protein] |
0.219 |
-0.054 |
| 56 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
0.218 |
-0.279 |
| 57 |
247024_at |
AT5G66985
|
unknown |
-0.998 |
-0.016 |
| 58 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
-0.955 |
-0.036 |
| 59 |
247070_at |
AT5G66815
|
unknown |
-0.945 |
0.005 |
| 60 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.908 |
-0.034 |
| 61 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-0.754 |
0.006 |
| 62 |
247212_at |
AT5G65040
|
unknown |
-0.742 |
-0.149 |
| 63 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.705 |
0.501 |
| 64 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
-0.684 |
-0.007 |
| 65 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.651 |
0.270 |
| 66 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.640 |
0.758 |
| 67 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
-0.606 |
-0.079 |
| 68 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.602 |
0.117 |
| 69 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.601 |
0.286 |
| 70 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.592 |
0.001 |
| 71 |
261221_at |
AT1G19960
|
unknown |
-0.572 |
-0.237 |
| 72 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.541 |
-0.060 |
| 73 |
250949_at |
AT5G03510
|
[C2H2-type zinc finger family protein] |
-0.533 |
-0.145 |
| 74 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.530 |
0.198 |
| 75 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
-0.529 |
0.180 |
| 76 |
252347_at |
AT3G48130
|
RSU1 |
-0.521 |
0.022 |
| 77 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
-0.517 |
0.160 |
| 78 |
257925_at |
AT3G23170
|
unknown |
-0.479 |
0.469 |
| 79 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.455 |
0.393 |
| 80 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.440 |
0.606 |
| 81 |
253919_at |
AT4G27350
|
unknown |
-0.431 |
0.028 |
| 82 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.429 |
0.489 |
| 83 |
245034_at |
AT2G26390
|
[Serine protease inhibitor (SERPIN) family protein] |
-0.412 |
0.335 |
| 84 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.410 |
-0.284 |
| 85 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
-0.409 |
-0.363 |
| 86 |
255659_at |
AT4G00895
|
[ATPase, F1 complex, OSCP/delta subunit protein] |
-0.401 |
-0.145 |
| 87 |
249870_at |
AT5G23080
|
TGH, TOUGH |
-0.391 |
-0.080 |
| 88 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.391 |
0.828 |
| 89 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
-0.387 |
0.033 |
| 90 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.375 |
0.162 |
| 91 |
248624_at |
AT5G48790
|
unknown |
-0.371 |
-0.121 |
| 92 |
264385_at |
AT1G12020
|
unknown |
-0.369 |
-0.275 |
| 93 |
257611_at |
AT3G26580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.358 |
0.065 |
| 94 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
-0.347 |
-0.003 |
| 95 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
-0.346 |
0.029 |
| 96 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.343 |
-0.073 |
| 97 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.341 |
0.833 |
| 98 |
256856_at |
AT3G15110
|
unknown |
-0.335 |
-0.004 |
| 99 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.334 |
0.009 |
| 100 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
-0.334 |
-0.061 |
| 101 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
-0.330 |
0.279 |
| 102 |
263688_at |
AT1G26920
|
unknown |
-0.329 |
0.164 |
| 103 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.326 |
0.107 |
| 104 |
256114_at |
AT1G16850
|
unknown |
-0.319 |
0.455 |
| 105 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.318 |
-0.079 |
| 106 |
254715_at |
AT4G13550
|
[triglyceride lipases] |
-0.318 |
-0.180 |
| 107 |
261508_at |
AT1G71730
|
unknown |
-0.315 |
-0.001 |
| 108 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.310 |
0.250 |
| 109 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.310 |
-0.391 |
| 110 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
-0.304 |
0.400 |
| 111 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
-0.301 |
0.505 |