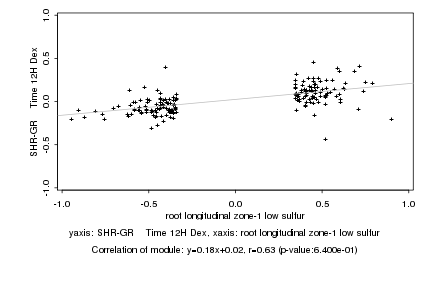
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root longitudinal zone-1 low sulfur |
Log2 signal ratio
SHR-GR Time 12H Dex |
| 1 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.900 |
-0.197 |
| 2 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.789 |
0.218 |
| 3 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.745 |
0.221 |
| 4 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
0.735 |
0.118 |
| 5 |
248057_at |
AT5G55520
|
unknown |
0.714 |
0.410 |
| 6 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.707 |
-0.091 |
| 7 |
245617_at |
AT4G14490
|
[SMAD/FHA domain-containing protein ] |
0.684 |
0.355 |
| 8 |
262081_at |
AT1G59540
|
ZCF125 |
0.631 |
0.215 |
| 9 |
262702_at |
AT1G16220
|
[Protein phosphatase 2C family protein] |
0.624 |
0.149 |
| 10 |
250586_at |
AT5G07630
|
[lipid transporters] |
0.623 |
0.151 |
| 11 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
0.603 |
-0.004 |
| 12 |
254156_at |
AT4G24490
|
ATRGTA1, RAB geranylgeranyl transferase alpha subunit 1, RGTA1, RAB geranylgeranyl transferase alpha subunit 1 |
0.602 |
0.028 |
| 13 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
0.600 |
0.354 |
| 14 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.595 |
0.091 |
| 15 |
251718_at |
AT3G56100
|
IMK3, MRLK, meristematic receptor-like kinase |
0.586 |
0.385 |
| 16 |
249430_at |
AT5G39900
|
[Small GTP-binding protein] |
0.583 |
0.068 |
| 17 |
265887_at |
AT2G25710
|
HCS1, holocarboxylase synthase 1 |
0.566 |
0.140 |
| 18 |
253969_at |
AT4G26430
|
CSN6B, COP9 signalosome subunit 6B |
0.555 |
0.246 |
| 19 |
248874_at |
AT5G46460
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.544 |
0.105 |
| 20 |
265011_at |
AT1G24490
|
ALB4, ALBINA 4, ARTEMIS, ARABIDOPSIS THALIANA ENVELOPE MEMBRANE INTEGRASE |
0.530 |
0.098 |
| 21 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.525 |
0.247 |
| 22 |
267224_at |
AT2G43990
|
unknown |
0.525 |
0.154 |
| 23 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.522 |
0.073 |
| 24 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
0.519 |
-0.437 |
| 25 |
266136_at |
AT2G45060
|
[Uncharacterised conserved protein UCP022280] |
0.518 |
0.048 |
| 26 |
259321_at |
AT3G01040
|
GAUT13, galacturonosyltransferase 13 |
0.518 |
-0.029 |
| 27 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
0.517 |
0.081 |
| 28 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
0.515 |
0.062 |
| 29 |
245869_at |
AT1G26330
|
[DNA binding] |
0.498 |
0.143 |
| 30 |
246415_at |
AT5G17160
|
unknown |
0.489 |
0.233 |
| 31 |
258951_at |
AT3G01380
|
[transferases] |
0.489 |
0.110 |
| 32 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.487 |
0.060 |
| 33 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
0.476 |
-0.005 |
| 34 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
0.476 |
0.270 |
| 35 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.471 |
0.172 |
| 36 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.462 |
0.030 |
| 37 |
250654_at |
AT5G06940
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.459 |
0.129 |
| 38 |
247667_at |
AT5G60150
|
unknown |
0.459 |
0.198 |
| 39 |
248131_at |
AT5G54830
|
[DOMON domain-containing protein / dopamine beta-monooxygenase N-terminal domain-containing protein] |
0.458 |
0.097 |
| 40 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
0.456 |
0.199 |
| 41 |
250531_at |
AT5G08650
|
[Small GTP-binding protein] |
0.456 |
0.163 |
| 42 |
247190_at |
AT5G65420
|
CYCD4;1, CYCLIN D4;1 |
0.455 |
-0.151 |
| 43 |
262693_at |
AT1G62780
|
unknown |
0.452 |
0.049 |
| 44 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
0.449 |
0.273 |
| 45 |
248498_at |
AT5G50390
|
EMB3141, EMBRYO DEFECTIVE 3141 |
0.448 |
0.051 |
| 46 |
266002_at |
AT2G37310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.448 |
0.232 |
| 47 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
0.447 |
0.463 |
| 48 |
263719_at |
AT2G13600
|
SLO2, SLOW GROWTH 2 |
0.446 |
-0.014 |
| 49 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
0.440 |
0.059 |
| 50 |
250309_at |
AT5G12220
|
[las1-like family protein] |
0.439 |
0.040 |
| 51 |
261208_at |
AT1G12930
|
[ARM repeat superfamily protein] |
0.439 |
0.126 |
| 52 |
264222_at |
AT1G60230
|
[Radical SAM superfamily protein] |
0.438 |
0.128 |
| 53 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.435 |
0.173 |
| 54 |
246458_at |
AT5G16860
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.434 |
0.118 |
| 55 |
248737_at |
AT5G48120
|
MET18, homolog of yeast MET18 |
0.434 |
0.139 |
| 56 |
246409_at |
AT1G57700
|
[Protein kinase superfamily protein] |
0.430 |
0.000 |
| 57 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
0.427 |
0.034 |
| 58 |
259411_at |
AT1G13410
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.425 |
0.130 |
| 59 |
247310_at |
AT5G63950
|
CHR24, chromatin remodeling 24 |
0.423 |
0.179 |
| 60 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
0.419 |
0.275 |
| 61 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.409 |
0.133 |
| 62 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.409 |
-0.004 |
| 63 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
0.408 |
0.073 |
| 64 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.406 |
0.094 |
| 65 |
245109_at |
AT2G41520
|
DJC62, DNA J protein C62, TPR15, tetratricopeptide repeat 15 |
0.406 |
0.134 |
| 66 |
266529_at |
AT2G16880
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.405 |
0.117 |
| 67 |
264259_at |
AT1G09290
|
unknown |
0.404 |
0.124 |
| 68 |
259600_at |
AT1G35220
|
unknown |
0.403 |
-0.053 |
| 69 |
264761_at |
AT1G61280
|
[Phosphatidylinositol N-acetylglucosaminyltransferase, GPI19/PIG-P subunit] |
0.400 |
-0.044 |
| 70 |
246734_at |
AT5G27680
|
RECQSIM, RECQ helicase SIM |
0.400 |
0.069 |
| 71 |
261463_at |
AT1G07740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.398 |
0.143 |
| 72 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
0.391 |
0.038 |
| 73 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.388 |
0.235 |
| 74 |
249388_at |
AT5G40090
|
CHL1, CHS1-like 1 |
0.385 |
0.186 |
| 75 |
266539_at |
AT2G35160
|
SGD9, SET DOMAIN-CONTAINING PROTEIN 9, SUVH5, SU(VAR)3-9 homolog 5 |
0.381 |
0.111 |
| 76 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.377 |
0.139 |
| 77 |
245972_at |
AT5G20680
|
TBL16, TRICHOME BIREFRINGENCE-LIKE 16 |
0.369 |
0.030 |
| 78 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
0.365 |
0.009 |
| 79 |
258174_at |
AT3G21470
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.363 |
0.082 |
| 80 |
267163_at |
AT2G37520
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.362 |
0.064 |
| 81 |
263415_at |
AT2G17250
|
EMB2762, EMBRYO DEFECTIVE 2762, NOC4, NucleOlar Complex associated 4 |
0.355 |
0.015 |
| 82 |
264228_at |
AT1G67490
|
GCS1, glucosidase 1, KNF, KNOPF |
0.354 |
0.063 |
| 83 |
246005_at |
AT5G08415
|
LIP1, lipoyl synthase 1 |
0.352 |
0.098 |
| 84 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.352 |
0.078 |
| 85 |
261604_at |
AT1G49590
|
ZOP1, Zinc-finger and OCRE domain-containing Protein 1 |
0.351 |
0.077 |
| 86 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
0.350 |
-0.104 |
| 87 |
262477_at |
AT1G11220
|
unknown |
0.348 |
0.316 |
| 88 |
247500_at |
AT5G61910
|
[DCD (Development and Cell Death) domain protein] |
0.345 |
0.044 |
| 89 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
0.345 |
0.153 |
| 90 |
254664_at |
AT4G18300
|
[Trimeric LpxA-like enzyme] |
0.342 |
0.201 |
| 91 |
252351_at |
AT3G48210
|
unknown |
0.342 |
0.163 |
| 92 |
246424_at |
AT5G17020
|
ATCRM1, ATXPO1, ARABIDOPSIS THALIANA EXPORTIN 1, HIT2, HEAT-INTOLERANT 2, XPO1, EXPORTIN 1, XPO1A, exportin 1A |
0.342 |
0.085 |
| 93 |
251680_at |
AT3G57060
|
[binding] |
0.341 |
0.247 |
| 94 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.950 |
-0.206 |
| 95 |
266259_at |
AT2G27830
|
unknown |
-0.911 |
-0.093 |
| 96 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
-0.876 |
-0.176 |
| 97 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.812 |
-0.107 |
| 98 |
261657_at |
AT1G50050
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.768 |
-0.150 |
| 99 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.756 |
-0.199 |
| 100 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
-0.708 |
-0.077 |
| 101 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.678 |
-0.056 |
| 102 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.628 |
-0.141 |
| 103 |
245906_at |
AT5G11070
|
unknown |
-0.620 |
-0.171 |
| 104 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
-0.612 |
0.139 |
| 105 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.609 |
-0.040 |
| 106 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
-0.603 |
-0.139 |
| 107 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
-0.592 |
-0.011 |
| 108 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.587 |
-0.099 |
| 109 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.586 |
-0.089 |
| 110 |
247061_at |
AT5G66780
|
unknown |
-0.580 |
-0.001 |
| 111 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.562 |
-0.102 |
| 112 |
261221_at |
AT1G19960
|
unknown |
-0.557 |
-0.068 |
| 113 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.556 |
-0.100 |
| 114 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.553 |
0.012 |
| 115 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.544 |
-0.128 |
| 116 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.542 |
-0.121 |
| 117 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
-0.529 |
0.174 |
| 118 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.523 |
-0.052 |
| 119 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.517 |
-0.127 |
| 120 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.516 |
-0.101 |
| 121 |
259479_at |
AT1G19020
|
unknown |
-0.516 |
-0.087 |
| 122 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.510 |
0.028 |
| 123 |
258306_at |
AT3G30530
|
ATBZIP42, basic leucine-zipper 42, bZIP42, basic leucine-zipper 42 |
-0.508 |
-0.001 |
| 124 |
258248_at |
AT3G29140
|
unknown |
-0.501 |
0.023 |
| 125 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.490 |
-0.311 |
| 126 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.488 |
-0.098 |
| 127 |
267226_at |
AT2G44010
|
unknown |
-0.488 |
-0.119 |
| 128 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
-0.483 |
-0.110 |
| 129 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.475 |
-0.156 |
| 130 |
253827_at |
AT4G28085
|
unknown |
-0.465 |
-0.180 |
| 131 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.464 |
-0.094 |
| 132 |
262539_at |
AT1G17200
|
[Uncharacterised protein family (UPF0497)] |
-0.460 |
-0.171 |
| 133 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.459 |
-0.121 |
| 134 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.458 |
-0.032 |
| 135 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
-0.453 |
-0.074 |
| 136 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.452 |
-0.270 |
| 137 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.451 |
0.133 |
| 138 |
262884_at |
AT1G64720
|
CP5 |
-0.439 |
-0.081 |
| 139 |
247407_at |
AT5G62900
|
unknown |
-0.436 |
-0.011 |
| 140 |
247851_at |
AT5G58070
|
ATTIL, TEMPERATURE-INDUCED LIPOCALIN, TIL, temperature-induced lipocalin |
-0.436 |
0.026 |
| 141 |
254200_at |
AT4G24110
|
unknown |
-0.434 |
-0.070 |
| 142 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.434 |
-0.046 |
| 143 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.433 |
0.096 |
| 144 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.433 |
0.038 |
| 145 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.430 |
-0.172 |
| 146 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.426 |
-0.032 |
| 147 |
265169_x_at |
AT1G23720
|
[Proline-rich extensin-like family protein] |
-0.422 |
-0.024 |
| 148 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
-0.420 |
0.015 |
| 149 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.419 |
-0.221 |
| 150 |
264772_at |
AT1G22930
|
[T-complex protein 11] |
-0.417 |
-0.064 |
| 151 |
247745_at |
AT5G59030
|
COPT1, copper transporter 1 |
-0.417 |
-0.039 |
| 152 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.408 |
0.398 |
| 153 |
262162_at |
AT1G78020
|
unknown |
-0.404 |
0.033 |
| 154 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
-0.401 |
-0.006 |
| 155 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.401 |
-0.158 |
| 156 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
-0.400 |
-0.003 |
| 157 |
251073_at |
AT5G01750
|
unknown |
-0.400 |
-0.076 |
| 158 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
-0.395 |
-0.013 |
| 159 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
-0.391 |
-0.018 |
| 160 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.387 |
-0.103 |
| 161 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
-0.386 |
0.033 |
| 162 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.385 |
-0.089 |
| 163 |
261026_at |
AT1G01240
|
unknown |
-0.383 |
-0.121 |
| 164 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.378 |
-0.021 |
| 165 |
247394_at |
AT5G62865
|
unknown |
-0.376 |
-0.175 |
| 166 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
-0.375 |
-0.120 |
| 167 |
262749_at |
AT1G28580
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.371 |
-0.101 |
| 168 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.369 |
-0.121 |
| 169 |
258263_at |
AT3G15780
|
unknown |
-0.367 |
-0.104 |
| 170 |
264082_at |
AT2G28570
|
unknown |
-0.363 |
-0.036 |
| 171 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
-0.363 |
0.016 |
| 172 |
245913_at |
AT5G19860
|
unknown |
-0.362 |
-0.131 |
| 173 |
253302_at |
AT4G33660
|
unknown |
-0.361 |
-0.117 |
| 174 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.360 |
-0.135 |
| 175 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.358 |
-0.196 |
| 176 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
-0.358 |
0.052 |
| 177 |
259340_at |
AT3G03870
|
unknown |
-0.353 |
-0.034 |
| 178 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
-0.350 |
-0.073 |
| 179 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
-0.349 |
0.018 |
| 180 |
263669_at |
AT1G04400
|
AT-PHH1, ATCRY2, CRY2, cryptochrome 2, FHA, PHH1 |
-0.347 |
-0.067 |
| 181 |
252976_s_at |
AT4G38550
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.346 |
-0.085 |
| 182 |
261551_at |
AT1G63440
|
HMA5, heavy metal atpase 5 |
-0.343 |
0.082 |
| 183 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
-0.342 |
0.027 |
| 184 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.341 |
-0.121 |
| 185 |
261083_at |
AT1G07310
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.341 |
0.045 |