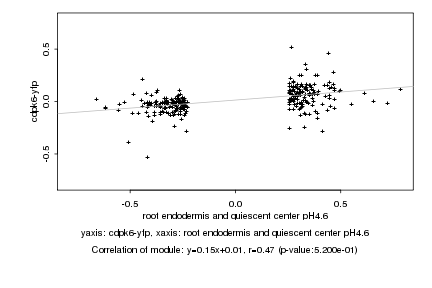
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root endodermis and quiescent center pH4.6 |
Log2 signal ratio
cdpk6-yfp |
| 1 |
247061_at |
AT5G66780
|
unknown |
0.782 |
0.119 |
| 2 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.719 |
-0.016 |
| 3 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.654 |
0.009 |
| 4 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.611 |
0.086 |
| 5 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.549 |
-0.019 |
| 6 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.495 |
0.112 |
| 7 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.492 |
0.101 |
| 8 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.470 |
0.025 |
| 9 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.470 |
0.113 |
| 10 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.467 |
0.132 |
| 11 |
252946_at |
AT4G39235
|
unknown |
0.466 |
-0.058 |
| 12 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.465 |
0.170 |
| 13 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.464 |
0.277 |
| 14 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.454 |
0.142 |
| 15 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.450 |
-0.044 |
| 16 |
251455_at |
AT3G60100
|
CSY5, citrate synthase 5 |
0.445 |
0.063 |
| 17 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.445 |
0.129 |
| 18 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.445 |
0.037 |
| 19 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.444 |
0.188 |
| 20 |
263515_at |
AT2G21640
|
unknown |
0.439 |
0.458 |
| 21 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.437 |
0.154 |
| 22 |
245076_at |
AT2G23170
|
GH3.3 |
0.436 |
-0.085 |
| 23 |
245352_at |
AT4G15490
|
UGT84A3 |
0.424 |
0.050 |
| 24 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.419 |
0.155 |
| 25 |
262578_at |
AT1G15300
|
CYTOSLEEPER, CYTOSLEEPER |
0.413 |
-0.026 |
| 26 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.411 |
-0.278 |
| 27 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.392 |
0.097 |
| 28 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.389 |
-0.161 |
| 29 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.389 |
0.070 |
| 30 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.388 |
0.256 |
| 31 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.386 |
-0.106 |
| 32 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.378 |
0.256 |
| 33 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.377 |
-0.088 |
| 34 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.374 |
0.165 |
| 35 |
256426_at |
AT1G33420
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.373 |
0.088 |
| 36 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.371 |
0.074 |
| 37 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.368 |
0.109 |
| 38 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.366 |
0.086 |
| 39 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.366 |
0.009 |
| 40 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
0.363 |
0.030 |
| 41 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.363 |
0.070 |
| 42 |
264024_at |
AT2G21180
|
unknown |
0.363 |
-0.037 |
| 43 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.361 |
0.117 |
| 44 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.358 |
0.137 |
| 45 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.356 |
0.067 |
| 46 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.355 |
0.158 |
| 47 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.352 |
0.067 |
| 48 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.351 |
0.065 |
| 49 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.350 |
0.161 |
| 50 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.350 |
0.071 |
| 51 |
256284_at |
AT3G12530
|
PSF2 |
0.349 |
0.083 |
| 52 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.349 |
0.069 |
| 53 |
256881_at |
AT3G26410
|
AtTRM11, TRM11, tRNA modification 11 |
0.349 |
0.083 |
| 54 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.347 |
-0.121 |
| 55 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.347 |
0.153 |
| 56 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.345 |
-0.011 |
| 57 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.341 |
-0.007 |
| 58 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.339 |
0.111 |
| 59 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.337 |
0.163 |
| 60 |
251564_at |
AT3G58160
|
ATMYOS3, ATXIJ, MYOSIN XI J, MYA3, MYOSIN A3, XI-16, MYOSIN XI-16, XIJ |
0.337 |
-0.000 |
| 61 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.337 |
0.309 |
| 62 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.333 |
0.140 |
| 63 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.332 |
0.130 |
| 64 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.332 |
0.011 |
| 65 |
261377_at |
AT1G18850
|
unknown |
0.331 |
0.116 |
| 66 |
260522_x_at |
AT2G41730
|
unknown |
0.330 |
0.354 |
| 67 |
259227_at |
AT3G07750
|
[3'-5'-exoribonuclease family protein] |
0.329 |
0.141 |
| 68 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.329 |
0.071 |
| 69 |
253305_at |
AT4G33666
|
unknown |
0.328 |
-0.119 |
| 70 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.325 |
0.046 |
| 71 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.324 |
-0.242 |
| 72 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.324 |
-0.107 |
| 73 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.321 |
0.016 |
| 74 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.317 |
0.151 |
| 75 |
260845_at |
AT1G17310
|
[MADS-box transcription factor family protein] |
0.317 |
-0.019 |
| 76 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.316 |
0.093 |
| 77 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.314 |
0.102 |
| 78 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.314 |
-0.038 |
| 79 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.313 |
0.256 |
| 80 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.312 |
0.167 |
| 81 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.312 |
-0.000 |
| 82 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.311 |
-0.050 |
| 83 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
0.309 |
-0.024 |
| 84 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.308 |
-0.064 |
| 85 |
259082_at |
AT3G04820
|
[Pseudouridine synthase family protein] |
0.308 |
0.097 |
| 86 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.307 |
0.062 |
| 87 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.306 |
0.150 |
| 88 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.306 |
-0.124 |
| 89 |
257352_at |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
0.304 |
-0.015 |
| 90 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.303 |
0.082 |
| 91 |
258202_at |
AT3G13940
|
[DNA binding] |
0.303 |
0.116 |
| 92 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.302 |
0.250 |
| 93 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.300 |
-0.070 |
| 94 |
246094_at |
AT5G19300
|
unknown |
0.295 |
0.065 |
| 95 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.293 |
0.171 |
| 96 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
0.292 |
0.023 |
| 97 |
263104_at |
AT2G05120
|
[Nucleoporin, Nup133/Nup155-like] |
0.291 |
-0.015 |
| 98 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.291 |
0.048 |
| 99 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.290 |
0.093 |
| 100 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.290 |
0.127 |
| 101 |
253659_at |
AT4G30150
|
unknown |
0.289 |
0.084 |
| 102 |
264027_at |
AT2G03670
|
CDC48B, cell division cycle 48B |
0.287 |
0.138 |
| 103 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.287 |
-0.044 |
| 104 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.285 |
0.151 |
| 105 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.283 |
0.066 |
| 106 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.281 |
0.029 |
| 107 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.281 |
0.036 |
| 108 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.280 |
0.024 |
| 109 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.279 |
0.100 |
| 110 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.278 |
-0.024 |
| 111 |
262219_at |
AT1G74750
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.275 |
0.052 |
| 112 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.275 |
0.197 |
| 113 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.274 |
0.149 |
| 114 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.273 |
0.187 |
| 115 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.273 |
0.121 |
| 116 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.273 |
0.138 |
| 117 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
0.271 |
0.007 |
| 118 |
263151_at |
AT1G54120
|
unknown |
0.271 |
-0.076 |
| 119 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.269 |
0.069 |
| 120 |
262943_at |
AT1G79470
|
[Aldolase-type TIM barrel family protein] |
0.268 |
0.107 |
| 121 |
263239_at |
AT2G16570
|
ASE1, GLN phosphoribosyl pyrophosphate amidotransferase 1, ATASE, ATASE1, GLN phosphoribosyl pyrophosphate amidotransferase 1 |
0.266 |
0.021 |
| 122 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.264 |
0.151 |
| 123 |
257611_at |
AT3G26580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.263 |
0.030 |
| 124 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.262 |
0.517 |
| 125 |
256204_at |
AT1G50840
|
POLGAMMA2, polymerase gamma 2, PolIA, polymerase I A |
0.262 |
0.016 |
| 126 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.261 |
0.021 |
| 127 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.261 |
0.105 |
| 128 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.261 |
0.079 |
| 129 |
246559_at |
AT5G15550
|
[Transducin/WD40 repeat-like superfamily protein] |
0.260 |
0.113 |
| 130 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.259 |
0.220 |
| 131 |
266618_at |
AT2G35480
|
unknown |
0.258 |
0.013 |
| 132 |
247575_at |
AT5G61030
|
GR-RBP3, glycine-rich RNA-binding protein 3 |
0.258 |
0.101 |
| 133 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.257 |
0.019 |
| 134 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.257 |
0.005 |
| 135 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.256 |
-0.009 |
| 136 |
254238_at |
AT4G23540
|
[ARM repeat superfamily protein] |
0.256 |
0.172 |
| 137 |
260049_at |
AT1G29940
|
NRPA2, nuclear RNA polymerase A2 |
0.255 |
0.107 |
| 138 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.255 |
-0.249 |
| 139 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.255 |
0.151 |
| 140 |
257033_at |
AT3G19170
|
ATPREP1, presequence protease 1, ATZNMP, PREP1, presequence protease 1 |
0.254 |
0.023 |
| 141 |
254455_at |
AT4G21140
|
unknown |
0.254 |
0.081 |
| 142 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.254 |
-0.023 |
| 143 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
0.254 |
-0.074 |
| 144 |
247676_at |
AT5G59980
|
AtRPP30, GAF1, GAMETOPHYTE DEFECTIVE 1, RPP30 |
0.254 |
0.094 |
| 145 |
265204_at |
AT2G36650
|
unknown |
-0.661 |
0.021 |
| 146 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.620 |
-0.055 |
| 147 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.618 |
-0.066 |
| 148 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
-0.559 |
-0.083 |
| 149 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.551 |
-0.027 |
| 150 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
-0.528 |
-0.003 |
| 151 |
252882_at |
AT4G39675
|
unknown |
-0.509 |
-0.384 |
| 152 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.492 |
-0.108 |
| 153 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.488 |
0.073 |
| 154 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.462 |
-0.112 |
| 155 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.447 |
0.017 |
| 156 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
-0.446 |
-0.041 |
| 157 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.444 |
0.214 |
| 158 |
257613_at |
AT3G26610
|
[Pectin lyase-like superfamily protein] |
-0.433 |
-0.015 |
| 159 |
258001_at |
AT3G28950
|
[AIG2-like (avirulence induced gene) family protein] |
-0.426 |
0.081 |
| 160 |
265134_at |
AT1G51260
|
LPAT3, lysophosphatidyl acyltransferase 3 |
-0.423 |
-0.103 |
| 161 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.420 |
-0.054 |
| 162 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.419 |
-0.009 |
| 163 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.419 |
-0.018 |
| 164 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.418 |
-0.528 |
| 165 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.413 |
-0.142 |
| 166 |
260628_at |
AT1G62320
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.413 |
-0.032 |
| 167 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.411 |
-0.018 |
| 168 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
-0.410 |
-0.004 |
| 169 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
-0.407 |
-0.034 |
| 170 |
256097_at |
AT1G13670
|
unknown |
-0.402 |
-0.010 |
| 171 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.400 |
0.062 |
| 172 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.398 |
-0.190 |
| 173 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.388 |
-0.097 |
| 174 |
262446_at |
AT1G49310
|
unknown |
-0.388 |
-0.131 |
| 175 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.385 |
-0.044 |
| 176 |
263616_at |
AT2G04680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.384 |
-0.009 |
| 177 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.382 |
-0.009 |
| 178 |
263915_at |
AT2G36430
|
unknown |
-0.378 |
0.087 |
| 179 |
256436_at |
AT3G11150
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.377 |
0.004 |
| 180 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.376 |
-0.045 |
| 181 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.375 |
-0.024 |
| 182 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.372 |
0.114 |
| 183 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.365 |
-0.030 |
| 184 |
262864_at |
AT1G64920
|
[UDP-Glycosyltransferase superfamily protein] |
-0.359 |
-0.051 |
| 185 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.359 |
-0.010 |
| 186 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.358 |
-0.119 |
| 187 |
267409_at |
AT2G34910
|
unknown |
-0.358 |
-0.040 |
| 188 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.356 |
-0.097 |
| 189 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.351 |
0.000 |
| 190 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.349 |
-0.015 |
| 191 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.349 |
-0.094 |
| 192 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.342 |
-0.099 |
| 193 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.341 |
-0.061 |
| 194 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.338 |
0.038 |
| 195 |
257502_at |
AT1G78110
|
unknown |
-0.337 |
-0.000 |
| 196 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.337 |
-0.059 |
| 197 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.337 |
-0.011 |
| 198 |
258648_at |
AT3G07900
|
[O-fucosyltransferase family protein] |
-0.335 |
0.001 |
| 199 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.335 |
-0.065 |
| 200 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.334 |
-0.053 |
| 201 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.332 |
-0.010 |
| 202 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.330 |
0.011 |
| 203 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.330 |
0.038 |
| 204 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.330 |
0.000 |
| 205 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.328 |
-0.099 |
| 206 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.327 |
-0.023 |
| 207 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.324 |
-0.009 |
| 208 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.318 |
-0.009 |
| 209 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.318 |
-0.111 |
| 210 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.314 |
-0.054 |
| 211 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.311 |
-0.133 |
| 212 |
259400_at |
AT1G17750
|
AtPEPR2, PEP1 RECEPTOR 2, PEPR2, PEP1 receptor 2 |
-0.310 |
-0.041 |
| 213 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
-0.306 |
0.026 |
| 214 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.303 |
-0.052 |
| 215 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.302 |
-0.098 |
| 216 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
-0.302 |
-0.056 |
| 217 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
-0.302 |
-0.005 |
| 218 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.300 |
-0.018 |
| 219 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.299 |
0.015 |
| 220 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
-0.299 |
-0.004 |
| 221 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.299 |
-0.006 |
| 222 |
255876_at |
AT2G40480
|
unknown |
-0.296 |
-0.128 |
| 223 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.296 |
-0.023 |
| 224 |
266640_at |
AT2G35585
|
unknown |
-0.295 |
0.004 |
| 225 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.292 |
-0.008 |
| 226 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.290 |
-0.122 |
| 227 |
263028_at |
AT1G24030
|
[Protein kinase superfamily protein] |
-0.290 |
-0.024 |
| 228 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.290 |
-0.235 |
| 229 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.287 |
0.029 |
| 230 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
-0.287 |
-0.029 |
| 231 |
254239_at |
AT4G23400
|
PIP1;5, plasma membrane intrinsic protein 1;5, PIP1D |
-0.286 |
-0.028 |
| 232 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.285 |
-0.089 |
| 233 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-0.285 |
-0.084 |
| 234 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.284 |
-0.066 |
| 235 |
257618_at |
AT3G24715
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.284 |
-0.028 |
| 236 |
264947_at |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.283 |
-0.042 |
| 237 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.282 |
-0.083 |
| 238 |
254264_at |
AT4G23510
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.281 |
0.006 |
| 239 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.277 |
-0.066 |
| 240 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.277 |
0.055 |
| 241 |
247945_at |
AT5G57150
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.275 |
0.026 |
| 242 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.273 |
0.061 |
| 243 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.271 |
-0.007 |
| 244 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.271 |
-0.022 |
| 245 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.271 |
0.032 |
| 246 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.271 |
-0.116 |
| 247 |
250828_at |
AT5G05250
|
unknown |
-0.270 |
0.111 |
| 248 |
252195_at |
AT3G50190
|
unknown |
-0.269 |
0.015 |
| 249 |
249484_at |
AT5G38970
|
ATBR6OX, BR6OX, BRASSINOSTEROID-6-OXIDASE, BR6OX1, brassinosteroid-6-oxidase 1, CYP85A1 |
-0.268 |
-0.004 |
| 250 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
-0.266 |
0.019 |
| 251 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
-0.265 |
-0.095 |
| 252 |
262609_at |
AT1G13930
|
unknown |
-0.264 |
0.068 |
| 253 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.262 |
0.003 |
| 254 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.262 |
-0.056 |
| 255 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.262 |
-0.067 |
| 256 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.261 |
-0.121 |
| 257 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.259 |
-0.020 |
| 258 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.258 |
-0.169 |
| 259 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.258 |
-0.060 |
| 260 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.257 |
-0.010 |
| 261 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.256 |
-0.061 |
| 262 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.256 |
0.008 |
| 263 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.254 |
0.036 |
| 264 |
253937_at |
AT4G26890
|
MAPKKK16, mitogen-activated protein kinase kinase kinase 16 |
-0.253 |
0.041 |
| 265 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.252 |
-0.022 |
| 266 |
262751_at |
AT1G16310
|
[Cation efflux family protein] |
-0.251 |
-0.043 |
| 267 |
259850_at |
AT1G72240
|
unknown |
-0.250 |
0.023 |
| 268 |
255448_at |
AT4G02810
|
FAF1, FANTASTIC FOUR 1 |
-0.250 |
-0.051 |
| 269 |
246135_at |
AT5G20885
|
[RING/U-box superfamily protein] |
-0.250 |
-0.073 |
| 270 |
254774_at |
AT4G13440
|
[Calcium-binding EF-hand family protein] |
-0.250 |
-0.029 |
| 271 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
-0.247 |
-0.046 |
| 272 |
264277_at |
AT1G60390
|
PG1, polygalacturonase 1 |
-0.246 |
-0.069 |
| 273 |
252211_at |
AT3G50220
|
IRX15, IRREGULAR XYLEM 15 |
-0.245 |
-0.032 |
| 274 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
-0.244 |
-0.004 |
| 275 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.244 |
0.029 |
| 276 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.242 |
-0.132 |
| 277 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
-0.242 |
0.012 |
| 278 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.241 |
-0.107 |
| 279 |
255104_at |
AT4G08685
|
SAH7 |
-0.240 |
-0.014 |
| 280 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
-0.239 |
-0.047 |
| 281 |
256154_at |
AT3G08490
|
unknown |
-0.237 |
-0.093 |
| 282 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.236 |
-0.283 |
| 283 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.234 |
-0.044 |
| 284 |
262315_at |
AT1G70990
|
[proline-rich family protein] |
-0.232 |
-0.050 |
| 285 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.230 |
-0.005 |
| 286 |
261278_at |
AT1G05800
|
DGL, DONGLE |
-0.230 |
-0.009 |
| 287 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.230 |
-0.052 |