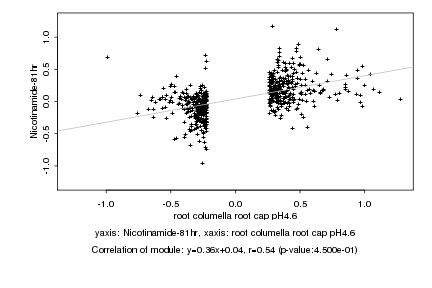
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root columella root cap pH4.6 |
Log2 signal ratio
Nicotinamide-81hr |
| 1 |
251871_at |
AT3G54520
|
unknown |
1.274 |
0.042 |
| 2 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
1.114 |
0.152 |
| 3 |
253305_at |
AT4G33666
|
unknown |
1.063 |
0.196 |
| 4 |
263515_at |
AT2G21640
|
unknown |
1.040 |
0.432 |
| 5 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.991 |
0.254 |
| 6 |
260522_x_at |
AT2G41730
|
unknown |
0.982 |
0.547 |
| 7 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.976 |
-0.072 |
| 8 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.962 |
-0.006 |
| 9 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.960 |
0.105 |
| 10 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.938 |
0.365 |
| 11 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.937 |
0.493 |
| 12 |
253831_at |
AT4G27580
|
unknown |
0.934 |
0.117 |
| 13 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.873 |
0.167 |
| 14 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.853 |
0.410 |
| 15 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.851 |
0.198 |
| 16 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.845 |
0.220 |
| 17 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.844 |
0.274 |
| 18 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.805 |
0.139 |
| 19 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.785 |
0.030 |
| 20 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.776 |
1.132 |
| 21 |
252864_at |
AT4G39740
|
HCC2, homologue of copper chaperone SCO1 2 |
0.771 |
0.110 |
| 22 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.738 |
0.435 |
| 23 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.724 |
0.064 |
| 24 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.710 |
0.317 |
| 25 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.706 |
0.664 |
| 26 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.677 |
0.174 |
| 27 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.674 |
0.190 |
| 28 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
0.673 |
0.179 |
| 29 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.654 |
0.146 |
| 30 |
265192_at |
AT1G05060
|
unknown |
0.652 |
0.127 |
| 31 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.643 |
0.250 |
| 32 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.636 |
0.814 |
| 33 |
264580_at |
AT1G05340
|
unknown |
0.626 |
0.438 |
| 34 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.608 |
-0.066 |
| 35 |
247061_at |
AT5G66780
|
unknown |
0.607 |
0.171 |
| 36 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.606 |
0.171 |
| 37 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
0.603 |
0.004 |
| 38 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.603 |
0.182 |
| 39 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.591 |
0.323 |
| 40 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.579 |
0.135 |
| 41 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.568 |
0.193 |
| 42 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.564 |
0.494 |
| 43 |
250823_at |
AT5G05180
|
unknown |
0.556 |
-0.389 |
| 44 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.550 |
0.186 |
| 45 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.549 |
0.011 |
| 46 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.539 |
0.355 |
| 47 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.535 |
0.117 |
| 48 |
266259_at |
AT2G27830
|
unknown |
0.535 |
0.264 |
| 49 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.525 |
0.562 |
| 50 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.523 |
-0.236 |
| 51 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.514 |
0.019 |
| 52 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.513 |
0.441 |
| 53 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.508 |
0.114 |
| 54 |
252946_at |
AT4G39235
|
unknown |
0.506 |
0.264 |
| 55 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
0.506 |
-0.195 |
| 56 |
249377_at |
AT5G40690
|
unknown |
0.505 |
0.145 |
| 57 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.502 |
0.344 |
| 58 |
253796_at |
AT4G28460
|
unknown |
0.501 |
0.695 |
| 59 |
253091_at |
AT4G37360
|
CYP81D2, cytochrome P450, family 81, subfamily D, polypeptide 2 |
0.498 |
0.137 |
| 60 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.497 |
0.579 |
| 61 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.494 |
0.561 |
| 62 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.491 |
0.589 |
| 63 |
248194_at |
AT5G54095
|
unknown |
0.488 |
0.032 |
| 64 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.485 |
-0.038 |
| 65 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.481 |
0.899 |
| 66 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.477 |
0.291 |
| 67 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.476 |
0.359 |
| 68 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
0.475 |
-0.098 |
| 69 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.474 |
0.371 |
| 70 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.469 |
0.453 |
| 71 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.468 |
0.833 |
| 72 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.468 |
0.786 |
| 73 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.465 |
-0.115 |
| 74 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
0.462 |
0.277 |
| 75 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.458 |
0.244 |
| 76 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.455 |
0.044 |
| 77 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.451 |
0.501 |
| 78 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.448 |
0.449 |
| 79 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.448 |
0.604 |
| 80 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
0.447 |
-0.002 |
| 81 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.446 |
0.017 |
| 82 |
263062_at |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.443 |
0.122 |
| 83 |
262219_at |
AT1G74750
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.443 |
0.189 |
| 84 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.441 |
0.068 |
| 85 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.440 |
0.231 |
| 86 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.438 |
0.669 |
| 87 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.438 |
-0.411 |
| 88 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.437 |
0.343 |
| 89 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.437 |
0.398 |
| 90 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.435 |
0.321 |
| 91 |
257670_at |
AT3G20340
|
unknown |
0.431 |
0.280 |
| 92 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.426 |
0.544 |
| 93 |
266587_at |
AT2G14880
|
[SWIB/MDM2 domain superfamily protein] |
0.424 |
0.037 |
| 94 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.423 |
0.395 |
| 95 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.420 |
0.128 |
| 96 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.418 |
0.602 |
| 97 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.415 |
0.036 |
| 98 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.414 |
-0.077 |
| 99 |
266618_at |
AT2G35480
|
unknown |
0.414 |
0.214 |
| 100 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.412 |
0.304 |
| 101 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.411 |
0.108 |
| 102 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.411 |
-0.029 |
| 103 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
0.410 |
0.289 |
| 104 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.409 |
0.393 |
| 105 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.408 |
0.219 |
| 106 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.408 |
0.330 |
| 107 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.407 |
0.335 |
| 108 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.407 |
-0.010 |
| 109 |
255716_at |
AT4G00330
|
CRCK2, calmodulin-binding receptor-like cytoplasmic kinase 2 |
0.407 |
0.082 |
| 110 |
253297_at |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
0.403 |
-0.128 |
| 111 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.401 |
0.179 |
| 112 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.398 |
0.178 |
| 113 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.398 |
0.408 |
| 114 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
0.398 |
0.269 |
| 115 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.397 |
-0.023 |
| 116 |
258571_at |
AT3G04420
|
anac048, NAC domain containing protein 48, NAC048, NAC domain containing protein 48 |
0.394 |
0.316 |
| 117 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.393 |
0.280 |
| 118 |
262578_at |
AT1G15300
|
CYTOSLEEPER, CYTOSLEEPER |
0.393 |
0.157 |
| 119 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.391 |
0.499 |
| 120 |
250152_at |
AT5G15120
|
unknown |
0.390 |
-0.107 |
| 121 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.390 |
0.071 |
| 122 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.388 |
0.495 |
| 123 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.386 |
0.478 |
| 124 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.384 |
0.136 |
| 125 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.384 |
-0.015 |
| 126 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.383 |
0.079 |
| 127 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.383 |
0.593 |
| 128 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.382 |
0.127 |
| 129 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.382 |
0.534 |
| 130 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.382 |
0.362 |
| 131 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.380 |
0.075 |
| 132 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.377 |
0.325 |
| 133 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.374 |
0.143 |
| 134 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.374 |
0.130 |
| 135 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
0.373 |
-0.021 |
| 136 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
0.373 |
0.048 |
| 137 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.372 |
0.133 |
| 138 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.372 |
0.317 |
| 139 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.371 |
0.248 |
| 140 |
263921_at |
AT2G36460
|
FBA6, fructose-bisphosphate aldolase 6 |
0.371 |
0.011 |
| 141 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.371 |
0.620 |
| 142 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.369 |
0.281 |
| 143 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.369 |
0.368 |
| 144 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.367 |
0.139 |
| 145 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.367 |
0.047 |
| 146 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.365 |
0.054 |
| 147 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
0.365 |
-0.025 |
| 148 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.364 |
0.155 |
| 149 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.362 |
0.483 |
| 150 |
257697_at |
AT3G12700
|
NANA, NANA |
0.360 |
0.128 |
| 151 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.359 |
0.272 |
| 152 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.357 |
0.160 |
| 153 |
266606_at |
AT2G46310
|
CRF5, cytokinin response factor 5 |
0.357 |
0.289 |
| 154 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.357 |
0.159 |
| 155 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.356 |
-0.206 |
| 156 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.356 |
0.173 |
| 157 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.355 |
-0.043 |
| 158 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.349 |
0.333 |
| 159 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.349 |
-0.062 |
| 160 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.348 |
-0.021 |
| 161 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.348 |
0.374 |
| 162 |
249750_at |
AT5G24570
|
unknown |
0.348 |
0.244 |
| 163 |
266004_at |
AT2G37330
|
ALS3, ALUMINUM SENSITIVE 3 |
0.344 |
0.005 |
| 164 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.344 |
0.200 |
| 165 |
247299_at |
AT5G63900
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.343 |
0.031 |
| 166 |
259465_at |
AT1G19030
|
unknown |
0.341 |
0.083 |
| 167 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
0.341 |
-0.257 |
| 168 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.340 |
0.227 |
| 169 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.340 |
0.134 |
| 170 |
250821_at |
AT5G05190
|
unknown |
0.339 |
0.363 |
| 171 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.339 |
0.824 |
| 172 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.338 |
0.227 |
| 173 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.337 |
0.247 |
| 174 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.337 |
0.265 |
| 175 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.337 |
0.216 |
| 176 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.337 |
0.314 |
| 177 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.336 |
0.773 |
| 178 |
252539_at |
AT3G45730
|
unknown |
0.334 |
0.706 |
| 179 |
249522_at |
AT5G38700
|
unknown |
0.334 |
0.131 |
| 180 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.332 |
0.532 |
| 181 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.332 |
0.652 |
| 182 |
264447_at |
AT1G27300
|
unknown |
0.332 |
0.256 |
| 183 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.331 |
0.572 |
| 184 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.330 |
0.063 |
| 185 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.330 |
0.087 |
| 186 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.330 |
0.215 |
| 187 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.329 |
-0.026 |
| 188 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.329 |
0.192 |
| 189 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.329 |
0.075 |
| 190 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.328 |
0.246 |
| 191 |
254018_at |
AT4G26180
|
CoAc2, CoA Carrier 2 |
0.327 |
0.025 |
| 192 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.327 |
-0.118 |
| 193 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.325 |
-0.087 |
| 194 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.324 |
0.041 |
| 195 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.324 |
0.139 |
| 196 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.322 |
0.173 |
| 197 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.321 |
0.058 |
| 198 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.320 |
0.458 |
| 199 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.320 |
0.078 |
| 200 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.320 |
-0.012 |
| 201 |
254263_at |
AT4G23493
|
unknown |
0.320 |
0.005 |
| 202 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.318 |
0.614 |
| 203 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.318 |
0.310 |
| 204 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.318 |
0.663 |
| 205 |
265850_at |
AT2G35720
|
OWL1, ORIENTATION UNDER VERY LOW FLUENCES OF LIGHT 1 |
0.315 |
0.216 |
| 206 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.314 |
0.013 |
| 207 |
247649_at |
AT5G60030
|
unknown |
0.314 |
-0.012 |
| 208 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.313 |
0.109 |
| 209 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.313 |
0.305 |
| 210 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.312 |
0.179 |
| 211 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.311 |
0.117 |
| 212 |
250225_at |
AT5G14105
|
unknown |
0.310 |
0.157 |
| 213 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.310 |
0.125 |
| 214 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.309 |
0.095 |
| 215 |
246125_at |
AT5G19875
|
unknown |
0.309 |
0.069 |
| 216 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.308 |
0.252 |
| 217 |
261239_at |
AT1G32930
|
AtGALT31A, GALT31A, glycosyltransferase of CAZY family GT31 A |
0.308 |
-0.208 |
| 218 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.307 |
-0.145 |
| 219 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.307 |
0.195 |
| 220 |
244901_at |
ATMG00640
|
ORF25 |
0.305 |
0.354 |
| 221 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.304 |
0.262 |
| 222 |
247989_at |
AT5G56350
|
[Pyruvate kinase family protein] |
0.304 |
0.154 |
| 223 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.301 |
0.195 |
| 224 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
0.301 |
-0.157 |
| 225 |
251776_at |
AT3G55620
|
eIF6A, eukaryotic initiation facor 6A, emb1624, embryo defective 1624 |
0.300 |
0.125 |
| 226 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.299 |
0.193 |
| 227 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.298 |
0.238 |
| 228 |
246305_at |
AT3G51890
|
CLC3, clathrin light chain 3 |
0.297 |
0.125 |
| 229 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.297 |
0.224 |
| 230 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.296 |
0.489 |
| 231 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.296 |
0.141 |
| 232 |
258451_at |
AT3G22360
|
AOX1B, alternative oxidase 1B |
0.295 |
0.209 |
| 233 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.294 |
-0.139 |
| 234 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.294 |
0.128 |
| 235 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.294 |
0.324 |
| 236 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.294 |
0.401 |
| 237 |
258201_at |
AT3G13910
|
unknown |
0.294 |
0.362 |
| 238 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.293 |
0.145 |
| 239 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.292 |
0.141 |
| 240 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.291 |
0.123 |
| 241 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.290 |
0.445 |
| 242 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.288 |
0.145 |
| 243 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.287 |
-0.146 |
| 244 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.287 |
0.398 |
| 245 |
246781_at |
AT5G27350
|
SFP1 |
0.287 |
0.006 |
| 246 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.287 |
0.242 |
| 247 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.285 |
0.085 |
| 248 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.285 |
0.023 |
| 249 |
263239_at |
AT2G16570
|
ASE1, GLN phosphoribosyl pyrophosphate amidotransferase 1, ATASE, ATASE1, GLN phosphoribosyl pyrophosphate amidotransferase 1 |
0.285 |
0.053 |
| 250 |
245776_at |
AT1G30260
|
unknown |
0.285 |
0.061 |
| 251 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.285 |
0.421 |
| 252 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.284 |
0.291 |
| 253 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.283 |
0.147 |
| 254 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.283 |
0.017 |
| 255 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.282 |
0.419 |
| 256 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.281 |
1.176 |
| 257 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.281 |
0.345 |
| 258 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.279 |
0.145 |
| 259 |
252683_at |
AT3G44380
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.279 |
0.147 |
| 260 |
246546_at |
AT5G15090
|
ATVDAC3, ARABIDOPSIS THALIANA VOLTAGE DEPENDENT ANION CHANNEL 3, VDAC3, voltage dependent anion channel 3 |
0.279 |
0.108 |
| 261 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.278 |
0.022 |
| 262 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.278 |
0.375 |
| 263 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.278 |
0.338 |
| 264 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.277 |
0.286 |
| 265 |
267279_at |
AT2G19460
|
unknown |
0.276 |
0.035 |
| 266 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.275 |
0.444 |
| 267 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.275 |
0.081 |
| 268 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
0.275 |
0.130 |
| 269 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.275 |
0.186 |
| 270 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.274 |
0.157 |
| 271 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.274 |
0.209 |
| 272 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.272 |
0.316 |
| 273 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
0.272 |
-0.134 |
| 274 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.272 |
0.398 |
| 275 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.271 |
0.318 |
| 276 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.271 |
0.112 |
| 277 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.271 |
0.127 |
| 278 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.269 |
-0.012 |
| 279 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.269 |
0.196 |
| 280 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.267 |
0.012 |
| 281 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.266 |
0.019 |
| 282 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.266 |
-0.077 |
| 283 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.266 |
0.433 |
| 284 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.265 |
-0.026 |
| 285 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.265 |
0.243 |
| 286 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.264 |
0.240 |
| 287 |
259275_at |
AT3G01060
|
unknown |
0.264 |
-0.001 |
| 288 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.264 |
0.305 |
| 289 |
259121_at |
AT3G02220
|
unknown |
0.264 |
-0.059 |
| 290 |
247277_at |
AT5G64420
|
[DNA polymerase V family] |
0.263 |
0.032 |
| 291 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.263 |
0.380 |
| 292 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
0.263 |
0.252 |
| 293 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.263 |
-0.018 |
| 294 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.262 |
0.306 |
| 295 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.261 |
-0.186 |
| 296 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.261 |
0.354 |
| 297 |
262584_at |
AT1G15440
|
ATPWP2, PERIODIC TRYPTOPHAN PROTEIN 2, PWP2, periodic tryptophan protein 2 |
0.260 |
0.235 |
| 298 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.260 |
0.144 |
| 299 |
260049_at |
AT1G29940
|
NRPA2, nuclear RNA polymerase A2 |
0.259 |
0.172 |
| 300 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.259 |
0.454 |
| 301 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.991 |
0.698 |
| 302 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.759 |
-0.174 |
| 303 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
-0.742 |
0.095 |
| 304 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.678 |
-0.123 |
| 305 |
245410_at |
AT4G17220
|
ATMAP70-5, microtubule-associated proteins 70-5, MAP70-5, microtubule-associated proteins 70-5 |
-0.655 |
0.018 |
| 306 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.648 |
0.077 |
| 307 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.638 |
-0.237 |
| 308 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.638 |
-0.112 |
| 309 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
-0.619 |
-0.013 |
| 310 |
262789_at |
AT1G10810
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.593 |
0.032 |
| 311 |
248356_at |
AT5G52350
|
ATEXO70A3, exocyst subunit exo70 family protein A3, EXO70A3, exocyst subunit exo70 family protein A3 |
-0.587 |
0.043 |
| 312 |
251125_at |
AT5G01060
|
BSK10, brassinosteroid-signaling kinase 10 |
-0.572 |
0.066 |
| 313 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.570 |
-0.106 |
| 314 |
261246_at |
AT1G20080
|
ATSYTB, NTMC2T1.2, NTMC2TYPE1.2, SYT2, synaptotagmin 2, SYTB |
-0.570 |
0.077 |
| 315 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-0.564 |
0.203 |
| 316 |
257637_at |
AT3G25810
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.543 |
0.042 |
| 317 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.542 |
-0.079 |
| 318 |
258740_at |
AT3G05780
|
LON3, lon protease 3 |
-0.541 |
0.098 |
| 319 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
-0.538 |
-0.252 |
| 320 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.514 |
-0.015 |
| 321 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.509 |
0.240 |
| 322 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.507 |
0.024 |
| 323 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.501 |
0.070 |
| 324 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.499 |
0.271 |
| 325 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.489 |
-0.003 |
| 326 |
265480_at |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
-0.488 |
-0.175 |
| 327 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.475 |
-0.579 |
| 328 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.474 |
0.237 |
| 329 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.474 |
0.150 |
| 330 |
264056_at |
AT2G28510
|
[Dof-type zinc finger DNA-binding family protein] |
-0.465 |
0.149 |
| 331 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.461 |
-0.573 |
| 332 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.461 |
0.389 |
| 333 |
266548_at |
AT2G35210
|
AGD10, MEE28, MATERNAL EFFECT EMBRYO ARREST 28, RPA, root and pollen arfgap |
-0.445 |
-0.045 |
| 334 |
259235_at |
AT3G11600
|
unknown |
-0.434 |
-0.024 |
| 335 |
259661_at |
AT1G55265
|
unknown |
-0.433 |
0.062 |
| 336 |
259464_at |
AT1G18990
|
unknown |
-0.430 |
0.088 |
| 337 |
253211_at |
AT4G34880
|
[Amidase family protein] |
-0.418 |
0.041 |
| 338 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.414 |
-0.104 |
| 339 |
263940_at |
AT2G35890
|
CPK25, calcium-dependent protein kinase 25 |
-0.413 |
-0.061 |
| 340 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.411 |
0.141 |
| 341 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.406 |
0.129 |
| 342 |
247905_at |
AT5G57400
|
unknown |
-0.400 |
0.065 |
| 343 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.398 |
-0.547 |
| 344 |
254781_at |
AT4G12840
|
unknown |
-0.397 |
-0.111 |
| 345 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.396 |
-0.319 |
| 346 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.393 |
-0.501 |
| 347 |
264278_at |
AT1G60130
|
[Mannose-binding lectin superfamily protein] |
-0.391 |
0.020 |
| 348 |
260523_at |
AT2G41720
|
EMB2654, EMBRYO DEFECTIVE 2654 |
-0.386 |
-0.031 |
| 349 |
247110_at |
AT5G65830
|
ATRLP57, receptor like protein 57, RLP57, receptor like protein 57 |
-0.385 |
0.126 |
| 350 |
258857_at |
AT3G02110
|
scpl25, serine carboxypeptidase-like 25 |
-0.382 |
0.059 |
| 351 |
260065_at |
AT1G73760
|
[RING/U-box superfamily protein] |
-0.381 |
0.059 |
| 352 |
248889_at |
AT5G46230
|
unknown |
-0.380 |
0.193 |
| 353 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.379 |
-0.174 |
| 354 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.378 |
-0.029 |
| 355 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.377 |
-0.045 |
| 356 |
250327_at |
AT5G12050
|
unknown |
-0.376 |
-0.165 |
| 357 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.375 |
-0.093 |
| 358 |
264896_at |
AT1G23210
|
AtGH9B6, glycosyl hydrolase 9B6, GH9B6, glycosyl hydrolase 9B6 |
-0.375 |
0.089 |
| 359 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.370 |
-0.155 |
| 360 |
258945_at |
AT3G10660
|
ATCPK2, CPK2, calmodulin-domain protein kinase cdpk isoform 2 |
-0.369 |
-0.120 |
| 361 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
-0.368 |
0.093 |
| 362 |
262759_at |
AT1G10800
|
unknown |
-0.367 |
0.004 |
| 363 |
261807_at |
AT1G30515
|
unknown |
-0.365 |
-0.016 |
| 364 |
263352_at |
AT2G22080
|
unknown |
-0.362 |
-0.429 |
| 365 |
251046_at |
AT5G02370
|
[ATP binding microtubule motor family protein] |
-0.359 |
-0.097 |
| 366 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.358 |
-0.246 |
| 367 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.358 |
-0.011 |
| 368 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
-0.355 |
-0.148 |
| 369 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.353 |
-0.668 |
| 370 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.352 |
0.069 |
| 371 |
253322_at |
AT4G33980
|
unknown |
-0.351 |
-0.086 |
| 372 |
248909_at |
AT5G45810
|
CIPK19, CBL-interacting protein kinase 19, SnRK3.5, SNF1-RELATED PROTEIN KINASE 3.5 |
-0.350 |
0.256 |
| 373 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.349 |
-0.452 |
| 374 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.347 |
-0.377 |
| 375 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.345 |
-0.442 |
| 376 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.344 |
-0.352 |
| 377 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
-0.344 |
0.044 |
| 378 |
253068_at |
AT4G37820
|
unknown |
-0.341 |
-0.226 |
| 379 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.337 |
0.032 |
| 380 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.337 |
-0.101 |
| 381 |
245602_at |
AT4G14270
|
unknown |
-0.333 |
-0.136 |
| 382 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.332 |
-0.046 |
| 383 |
261846_at |
AT1G11540
|
[Sulfite exporter TauE/SafE family protein] |
-0.332 |
0.087 |
| 384 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
-0.325 |
-0.006 |
| 385 |
260365_at |
AT1G70630
|
RAY1, REDUCED ARABINOSE YARIV 1 |
-0.324 |
0.006 |
| 386 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.324 |
-0.429 |
| 387 |
261877_at |
AT1G50580
|
[UDP-Glycosyltransferase superfamily protein] |
-0.323 |
0.043 |
| 388 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
-0.322 |
-0.175 |
| 389 |
246188_at |
AT5G21050
|
[LOCATED IN: chloroplast] |
-0.321 |
0.051 |
| 390 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.318 |
-0.064 |
| 391 |
264092_at |
AT1G79040
|
PSBR, photosystem II subunit R |
-0.318 |
0.052 |
| 392 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.316 |
-0.237 |
| 393 |
261657_at |
AT1G50050
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.316 |
-0.045 |
| 394 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.316 |
-0.048 |
| 395 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.315 |
-0.344 |
| 396 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.314 |
-0.251 |
| 397 |
247885_at |
AT5G57830
|
unknown |
-0.314 |
-0.210 |
| 398 |
267170_at |
AT2G37585
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.312 |
-0.222 |
| 399 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.309 |
-0.353 |
| 400 |
264714_at |
AT1G69990
|
[Leucine-rich repeat protein kinase family protein] |
-0.308 |
-0.009 |
| 401 |
249919_at |
AT5G19250
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.308 |
-0.033 |
| 402 |
247610_at |
AT5G60630
|
unknown |
-0.308 |
0.088 |
| 403 |
245245_at |
AT1G44318
|
hemb2 |
-0.308 |
0.064 |
| 404 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.308 |
-0.091 |
| 405 |
265183_at |
AT1G23750
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.307 |
-0.417 |
| 406 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.307 |
0.125 |
| 407 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.306 |
-0.309 |
| 408 |
260723_at |
AT1G48070
|
[Thioredoxin superfamily protein] |
-0.306 |
0.120 |
| 409 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.305 |
-0.043 |
| 410 |
250734_at |
AT5G06270
|
unknown |
-0.303 |
-0.408 |
| 411 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.301 |
-0.101 |
| 412 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.300 |
-0.395 |
| 413 |
256025_at |
AT1G58370
|
ATXYN1, ARABIDOPSIS THALIANA XYLANASE 1, RXF12 |
-0.300 |
-0.111 |
| 414 |
253855_at |
AT4G28050
|
TET7, tetraspanin7 |
-0.300 |
-0.301 |
| 415 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.300 |
-0.155 |
| 416 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.300 |
-0.055 |
| 417 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
-0.298 |
-0.227 |
| 418 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.298 |
-0.153 |
| 419 |
265790_at |
AT2G01170
|
AtGABP, BAT1, bidirectional amino acid transporter 1, GABP, GABA permease |
-0.298 |
-0.189 |
| 420 |
256660_at |
AT3G12060
|
TBL1, TRICHOME BIREFRINGENCE-LIKE 1 |
-0.297 |
-0.013 |
| 421 |
252191_at |
AT3G50180
|
unknown |
-0.297 |
0.084 |
| 422 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.297 |
-0.132 |
| 423 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
-0.296 |
-0.250 |
| 424 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.296 |
-0.497 |
| 425 |
266334_at |
AT2G32380
|
[Transmembrane protein 97, predicted] |
-0.296 |
-0.075 |
| 426 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.296 |
-0.215 |
| 427 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.294 |
-0.072 |
| 428 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.294 |
-0.196 |
| 429 |
261142_at |
AT1G19780
|
ATCNGC8, cyclic nucleotide gated channel 8, CNGC8, cyclic nucleotide gated channel 8 |
-0.293 |
0.037 |
| 430 |
255901_at |
AT1G17890
|
GER2 |
-0.293 |
-0.078 |
| 431 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.292 |
-0.053 |
| 432 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
-0.292 |
-0.121 |
| 433 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.291 |
-0.171 |
| 434 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
-0.291 |
-0.238 |
| 435 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.291 |
-0.067 |
| 436 |
247966_at |
AT5G56610
|
[Phosphotyrosine protein phosphatases superfamily protein] |
-0.289 |
-0.157 |
| 437 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
-0.289 |
0.006 |
| 438 |
251854_at |
AT3G54800
|
[Pleckstrin homology (PH) and lipid-binding START domains-containing protein] |
-0.289 |
0.096 |
| 439 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.288 |
-0.277 |
| 440 |
264587_at |
AT1G05200
|
ATGLR3.4, glutamate receptor 3.4, GLR3.4, glutamate receptor 3.4, GLUR3 |
-0.287 |
0.030 |
| 441 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.284 |
-0.198 |
| 442 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.283 |
-0.610 |
| 443 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.283 |
0.040 |
| 444 |
248548_at |
AT5G50300
|
ATAZG2, ARABIDOPSIS THALIANA AZA-GUANINE RESISTANT2, AZG2, AZA-GUANINE RESISTANT2 |
-0.283 |
0.038 |
| 445 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.283 |
-0.177 |
| 446 |
259611_at |
AT1G52280
|
AtRABG3d, RAB GTPase homolog G3D, RABG3d, RAB GTPase homolog G3D |
-0.282 |
-0.149 |
| 447 |
266869_at |
AT2G44660
|
[ALG6, ALG8 glycosyltransferase family] |
-0.280 |
0.055 |
| 448 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.280 |
-0.299 |
| 449 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.280 |
-0.130 |
| 450 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.279 |
0.142 |
| 451 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.278 |
-0.277 |
| 452 |
245698_at |
AT5G04160
|
[Nucleotide-sugar transporter family protein] |
-0.278 |
-0.112 |
| 453 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.277 |
-0.381 |
| 454 |
249574_at |
AT5G37660
|
PDLP7, plasmodesmata-located protein 7 |
-0.276 |
0.207 |
| 455 |
257095_at |
AT3G20560
|
ATPDI12, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 12, ATPDIL5-3, PDI-like 5-3, PDI12, PROTEIN DISULFIDE ISOMERASE 12, PDIL5-3, PDI-like 5-3 |
-0.276 |
-0.038 |
| 456 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
-0.276 |
-0.110 |
| 457 |
254662_at |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
-0.274 |
-0.167 |
| 458 |
262200_at |
AT2G01070
|
[Lung seven transmembrane receptor family protein] |
-0.273 |
-0.083 |
| 459 |
260674_at |
AT1G19370
|
unknown |
-0.273 |
-0.078 |
| 460 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
-0.272 |
0.242 |
| 461 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.271 |
-0.066 |
| 462 |
250930_at |
AT5G03160
|
ATP58IPK, homolog of mamallian P58IPK, P58IPK, homolog of mamallian P58IPK |
-0.270 |
0.173 |
| 463 |
260644_at |
AT1G53290
|
[Galactosyltransferase family protein] |
-0.270 |
-0.099 |
| 464 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.270 |
0.139 |
| 465 |
253529_at |
AT4G31520
|
[SDA1 family protein] |
-0.269 |
0.010 |
| 466 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.269 |
-0.209 |
| 467 |
257647_at |
AT3G25805
|
unknown |
-0.268 |
-0.038 |
| 468 |
248407_at |
AT5G51500
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.268 |
-0.152 |
| 469 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
-0.267 |
-0.014 |
| 470 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.267 |
-0.439 |
| 471 |
252165_at |
AT3G50550
|
unknown |
-0.266 |
-0.069 |
| 472 |
248139_at |
AT5G54970
|
unknown |
-0.266 |
-0.175 |
| 473 |
247709_at |
AT5G59250
|
[Major facilitator superfamily protein] |
-0.266 |
-0.082 |
| 474 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
-0.265 |
-0.078 |
| 475 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.263 |
-0.076 |
| 476 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.263 |
-0.262 |
| 477 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.263 |
-0.314 |
| 478 |
265451_at |
AT2G46490
|
unknown |
-0.262 |
0.006 |
| 479 |
255332_at |
AT4G04340
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.262 |
-0.298 |
| 480 |
251076_at |
AT5G01970
|
unknown |
-0.261 |
0.046 |
| 481 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.260 |
0.001 |
| 482 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.260 |
-0.178 |
| 483 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
-0.260 |
-0.099 |
| 484 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.259 |
-0.955 |
| 485 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.259 |
-0.308 |
| 486 |
251660_at |
AT3G57160
|
unknown |
-0.259 |
-0.018 |
| 487 |
245685_at |
AT5G22220
|
ATE2FB, E2F1, E2F transcription factor 1, E2FB |
-0.259 |
0.257 |
| 488 |
252266_at |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.258 |
0.018 |
| 489 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.258 |
-0.297 |
| 490 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.257 |
-0.071 |
| 491 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.257 |
-0.077 |
| 492 |
245619_at |
AT4G13990
|
[Exostosin family protein] |
-0.256 |
-0.101 |
| 493 |
266926_at |
AT2G46000
|
unknown |
-0.255 |
-0.058 |
| 494 |
248359_at |
AT5G52410
|
unknown |
-0.255 |
0.028 |
| 495 |
259963_at |
AT1G53660
|
[nodulin MtN21 /EamA-like transporter family protein] |
-0.255 |
-0.047 |
| 496 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
-0.255 |
0.084 |
| 497 |
261639_at |
AT1G50010
|
TUA2, tubulin alpha-2 chain |
-0.254 |
-0.014 |
| 498 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
-0.254 |
-0.332 |
| 499 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.253 |
-0.118 |
| 500 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
-0.253 |
-0.165 |
| 501 |
259768_at |
AT1G29390
|
COR314-TM2, cold regulated 314 thylakoid membrane 2, COR413IM2, COLD REGULATED 314 INNER MEMBRANE 2 |
-0.253 |
-0.111 |
| 502 |
263762_at |
AT2G21380
|
[Kinesin motor family protein] |
-0.253 |
-0.327 |
| 503 |
258852_at |
AT3G06300
|
AT-P4H-2, P4H isoform 2, P4H2, prolyl 4-hydroxylase 2 |
-0.252 |
-0.102 |
| 504 |
265936_at |
AT2G19600
|
ATKEA4, K+ efflux antiporter 4, KEA4, KEA4, K+ efflux antiporter 4 |
-0.252 |
-0.147 |
| 505 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
-0.252 |
-0.172 |
| 506 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.252 |
-0.018 |
| 507 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
-0.252 |
-0.134 |
| 508 |
248131_at |
AT5G54830
|
[DOMON domain-containing protein / dopamine beta-monooxygenase N-terminal domain-containing protein] |
-0.251 |
0.135 |
| 509 |
266774_at |
AT2G29050
|
ATRBL1, RHOMBOID-like 1, RBL1, RHOMBOID-like 1 |
-0.251 |
-0.054 |
| 510 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.251 |
-0.478 |
| 511 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.251 |
-0.554 |
| 512 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
-0.249 |
-0.492 |
| 513 |
249334_at |
AT5G41000
|
AtYSL4, YSL4, YELLOW STRIPE like 4 |
-0.249 |
-0.053 |
| 514 |
247666_at |
AT5G60140
|
[AP2/B3-like transcriptional factor family protein] |
-0.249 |
0.025 |
| 515 |
247746_at |
AT5G58970
|
ATUCP2, uncoupling protein 2, UCP2, uncoupling protein 2 |
-0.249 |
-0.209 |
| 516 |
258976_at |
AT3G01980
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.249 |
-0.288 |
| 517 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.248 |
-0.268 |
| 518 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.248 |
0.102 |
| 519 |
250586_at |
AT5G07630
|
[lipid transporters] |
-0.248 |
-0.147 |
| 520 |
255490_at |
AT4G02660
|
[Beige/BEACH domain ] |
-0.248 |
0.039 |
| 521 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.248 |
0.066 |
| 522 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.247 |
-0.189 |
| 523 |
261505_at |
AT1G71696
|
SOL1, SUPPRESSOR OF LLP1 1 |
-0.247 |
-0.160 |
| 524 |
252608_at |
AT3G45090
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.246 |
-0.008 |
| 525 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.246 |
-0.360 |
| 526 |
256908_at |
AT3G24040
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.246 |
-0.066 |
| 527 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.246 |
-0.635 |
| 528 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
-0.245 |
0.003 |
| 529 |
248664_at |
AT5G48580
|
FKBP15-2, FK506- and rapamycin-binding protein 15 kD-2 |
-0.245 |
-0.116 |
| 530 |
251796_at |
AT3G55360
|
ATTSC13, CER10, ECERIFERUM 10, ECR, ENOYL-COA REDUCTASE, TSC13 |
-0.244 |
-0.087 |
| 531 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-0.243 |
-0.151 |
| 532 |
246511_at |
AT5G15490
|
UGD3, UDP-glucose dehydrogenase 3 |
-0.242 |
0.020 |
| 533 |
252879_at |
AT4G39390
|
ATNST-KT1, A. THALIANA NUCLEOTIDE SUGAR TRANSPORTER-KT 1, NST-K1, nucleotide sugar transporter-KT 1 |
-0.242 |
-0.078 |
| 534 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.242 |
-0.374 |
| 535 |
254077_at |
AT4G25640
|
ATDTX35, detoxifying efflux carrier 35, DTX35, detoxifying efflux carrier 35, FFT, FLOWER FLAVONOID TRANSPORTER |
-0.241 |
0.225 |
| 536 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.241 |
-0.145 |
| 537 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
-0.240 |
0.134 |
| 538 |
254659_at |
AT4G18240
|
ATSS4, ARABIDOPSIS THALIANA STARCH SYNTHASE 4, SS4, starch synthase 4, SSIV, STARCH SYNTHASE 4 |
-0.239 |
-0.270 |
| 539 |
259721_at |
AT1G60890
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
-0.238 |
0.154 |
| 540 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.238 |
0.094 |
| 541 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.238 |
-0.319 |
| 542 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.238 |
-0.186 |
| 543 |
262693_at |
AT1G62780
|
unknown |
-0.238 |
-0.162 |
| 544 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.237 |
-0.119 |
| 545 |
248001_at |
AT5G55990
|
ATCBL2, CBL2, calcineurin B-like protein 2 |
-0.237 |
-0.039 |
| 546 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.237 |
0.060 |
| 547 |
247881_at |
AT5G57700
|
[BNR/Asp-box repeat family protein] |
-0.236 |
-0.376 |
| 548 |
252352_at |
AT3G48185
|
unknown |
-0.236 |
0.017 |
| 549 |
264055_at |
AT2G28590
|
[Protein kinase superfamily protein] |
-0.236 |
-0.093 |
| 550 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.235 |
-0.699 |
| 551 |
245209_at |
AT5G12340
|
unknown |
-0.235 |
0.527 |
| 552 |
245423_at |
AT4G17483
|
[alpha/beta-Hydrolases superfamily protein] |
-0.234 |
0.105 |
| 553 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.234 |
0.077 |
| 554 |
261791_at |
AT1G16170
|
unknown |
-0.234 |
0.022 |
| 555 |
246772_at |
AT5G27490
|
[Integral membrane Yip1 family protein] |
-0.233 |
-0.135 |
| 556 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.233 |
0.717 |
| 557 |
246505_at |
AT5G16250
|
unknown |
-0.233 |
-0.361 |
| 558 |
249220_at |
AT5G42420
|
[Nucleotide-sugar transporter family protein] |
-0.233 |
-0.124 |
| 559 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
-0.233 |
0.044 |
| 560 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.233 |
-0.058 |
| 561 |
263619_at |
AT2G04650
|
[ADP-glucose pyrophosphorylase family protein] |
-0.233 |
-0.411 |
| 562 |
264581_at |
AT1G05210
|
[Transmembrane protein 97, predicted] |
-0.232 |
-0.173 |
| 563 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.232 |
-0.307 |
| 564 |
262775_at |
AT1G13000
|
unknown |
-0.231 |
-0.085 |
| 565 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.231 |
-0.156 |
| 566 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
-0.231 |
0.124 |
| 567 |
262504_at |
AT1G21750
|
ATPDI5, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 5, ATPDIL1-1, PDI-like 1-1, PDI5, PROTEIN DISULFIDE ISOMERASE 5, PDIL1-1, PDI-like 1-1 |
-0.231 |
0.022 |
| 568 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.231 |
-0.160 |
| 569 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.230 |
-0.351 |
| 570 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
-0.230 |
-0.261 |
| 571 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
-0.230 |
-0.103 |
| 572 |
261074_at |
AT1G07290
|
GONST2, golgi nucleotide sugar transporter 2 |
-0.230 |
0.032 |
| 573 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.230 |
-0.278 |
| 574 |
245293_at |
AT4G16660
|
[heat shock protein 70 (Hsp 70) family protein] |
-0.229 |
-0.065 |
| 575 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.229 |
-0.133 |
| 576 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.229 |
0.010 |
| 577 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.229 |
-0.408 |
| 578 |
262797_at |
AT1G20840
|
AtTMT1, TMT1, tonoplast monosaccharide transporter1 |
-0.229 |
0.064 |
| 579 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
-0.228 |
0.106 |
| 580 |
256913_at |
AT3G23870
|
unknown |
-0.228 |
-0.182 |
| 581 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.227 |
-0.224 |
| 582 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.227 |
0.629 |
| 583 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.227 |
-0.740 |
| 584 |
254548_at |
AT4G19865
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.226 |
-0.033 |
| 585 |
255948_at |
AT1G22060
|
[LOCATED IN: vacuole] |
-0.226 |
-0.304 |
| 586 |
258988_at |
AT3G08890
|
unknown |
-0.226 |
0.080 |
| 587 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.226 |
-0.316 |
| 588 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
-0.226 |
-0.312 |
| 589 |
256952_at |
AT3G21160
|
MANIA, ALPHA-MANNOSIDASE IA, MNS2, alpha-mannosidase 2 |
-0.225 |
0.090 |
| 590 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.225 |
-0.452 |
| 591 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.225 |
0.022 |
| 592 |
263579_at |
AT2G17030
|
unknown |
-0.225 |
0.167 |
| 593 |
261944_at |
AT1G64650
|
[Major facilitator superfamily protein] |
-0.225 |
-0.054 |
| 594 |
267544_at |
AT2G32720
|
ATCB5-B, ARABIDOPSIS CYTOCHROME B5 ISOFORM B, B5 #4, CB5-B, cytochrome B5 isoform B, CYTB5-D |
-0.225 |
-0.083 |
| 595 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-0.224 |
-0.324 |
| 596 |
261649_at |
AT1G27700
|
[Syntaxin/t-SNARE family protein] |
-0.224 |
-0.033 |
| 597 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.224 |
-0.317 |
| 598 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.224 |
0.089 |
| 599 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
-0.224 |
-0.167 |
| 600 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
-0.224 |
0.134 |