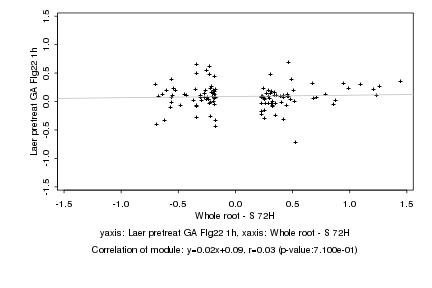
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Whole root - S 72H |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.441 |
0.366 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.255 |
0.276 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
1.229 |
0.110 |
| 4 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
1.206 |
0.215 |
| 5 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
1.088 |
0.302 |
| 6 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.986 |
0.233 |
| 7 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.945 |
0.326 |
| 8 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.871 |
0.032 |
| 9 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.858 |
-0.044 |
| 10 |
257421_at |
AT1G12030
|
unknown |
0.780 |
0.140 |
| 11 |
255105_at |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
0.705 |
0.072 |
| 12 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.676 |
0.054 |
| 13 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.670 |
0.321 |
| 14 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.517 |
-0.708 |
| 15 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
0.514 |
0.004 |
| 16 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.501 |
0.199 |
| 17 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.484 |
0.403 |
| 18 |
253525_at |
AT4G31330
|
unknown |
0.479 |
0.042 |
| 19 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.461 |
0.703 |
| 20 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
0.460 |
0.099 |
| 21 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.453 |
0.125 |
| 22 |
247061_at |
AT5G66780
|
unknown |
0.447 |
0.098 |
| 23 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
0.439 |
-0.063 |
| 24 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.417 |
0.119 |
| 25 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.416 |
-0.302 |
| 26 |
256584_at |
AT3G28750
|
unknown |
0.415 |
0.088 |
| 27 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.402 |
-0.002 |
| 28 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.388 |
0.089 |
| 29 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.356 |
0.106 |
| 30 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.346 |
-0.231 |
| 31 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.342 |
-0.029 |
| 32 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.341 |
0.118 |
| 33 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.334 |
0.163 |
| 34 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
0.324 |
0.008 |
| 35 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.322 |
-0.066 |
| 36 |
253317_at |
AT4G33960
|
unknown |
0.318 |
-0.087 |
| 37 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.313 |
0.180 |
| 38 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.313 |
-0.034 |
| 39 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.306 |
0.144 |
| 40 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.305 |
0.486 |
| 41 |
257595_at |
AT3G24750
|
unknown |
0.291 |
0.059 |
| 42 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.283 |
0.197 |
| 43 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
0.281 |
0.101 |
| 44 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.281 |
-0.030 |
| 45 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.271 |
0.147 |
| 46 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.255 |
-0.019 |
| 47 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.254 |
-0.285 |
| 48 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.252 |
0.041 |
| 49 |
246584_at |
AT5G14730
|
unknown |
0.247 |
0.063 |
| 50 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.247 |
-0.147 |
| 51 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.240 |
0.232 |
| 52 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
0.240 |
0.079 |
| 53 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.235 |
0.089 |
| 54 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.228 |
0.103 |
| 55 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.226 |
-0.224 |
| 56 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.225 |
-0.174 |
| 57 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.225 |
0.075 |
| 58 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.222 |
-0.033 |
| 59 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.702 |
0.308 |
| 60 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.692 |
-0.395 |
| 61 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.678 |
0.094 |
| 62 |
265611_at |
AT2G25510
|
unknown |
-0.639 |
0.125 |
| 63 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.621 |
-0.317 |
| 64 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.606 |
0.206 |
| 65 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.570 |
-0.090 |
| 66 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.566 |
0.079 |
| 67 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.565 |
0.388 |
| 68 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.564 |
-0.016 |
| 69 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.555 |
0.118 |
| 70 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.548 |
0.233 |
| 71 |
249112_at |
AT5G43780
|
APS4 |
-0.531 |
0.204 |
| 72 |
258362_at |
AT3G14280
|
unknown |
-0.482 |
-0.067 |
| 73 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.449 |
0.130 |
| 74 |
260495_at |
AT2G41810
|
unknown |
-0.435 |
0.117 |
| 75 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.374 |
0.035 |
| 76 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.350 |
0.228 |
| 77 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.347 |
0.655 |
| 78 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.347 |
0.494 |
| 79 |
252882_at |
AT4G39675
|
unknown |
-0.344 |
-0.075 |
| 80 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
-0.341 |
-0.264 |
| 81 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.341 |
-0.055 |
| 82 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.314 |
0.119 |
| 83 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.312 |
0.085 |
| 84 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.301 |
0.032 |
| 85 |
260744_at |
AT1G15010
|
unknown |
-0.276 |
0.154 |
| 86 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.273 |
0.071 |
| 87 |
257670_at |
AT3G20340
|
unknown |
-0.264 |
0.209 |
| 88 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.261 |
0.040 |
| 89 |
265945_at |
AT2G19500
|
ATCKX2, CKX2, cytokinin oxidase 2 |
-0.256 |
0.561 |
| 90 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.245 |
0.088 |
| 91 |
253038_at |
AT4G37790
|
HAT22 |
-0.240 |
0.047 |
| 92 |
265387_at |
AT2G20670
|
unknown |
-0.238 |
0.046 |
| 93 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.231 |
-0.022 |
| 94 |
245076_at |
AT2G23170
|
GH3.3 |
-0.229 |
0.484 |
| 95 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-0.228 |
0.624 |
| 96 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.226 |
0.230 |
| 97 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
-0.224 |
-0.248 |
| 98 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.219 |
-0.012 |
| 99 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
-0.215 |
0.164 |
| 100 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.210 |
0.269 |
| 101 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.207 |
0.094 |
| 102 |
258225_at |
AT3G15630
|
unknown |
-0.198 |
0.147 |
| 103 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.197 |
0.007 |
| 104 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.194 |
0.158 |
| 105 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.194 |
0.209 |
| 106 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
-0.191 |
0.442 |
| 107 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.190 |
-0.040 |
| 108 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.189 |
0.131 |
| 109 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.188 |
0.185 |
| 110 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.185 |
0.069 |
| 111 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.183 |
0.075 |
| 112 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.182 |
0.086 |
| 113 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.179 |
0.214 |
| 114 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
-0.175 |
-0.323 |
| 115 |
251310_at |
AT3G61150
|
HD-GL2-1, HOMEODOMAIN-GLABRA2 1, HDG1, homeodomain GLABROUS 1 |
-0.175 |
-0.437 |