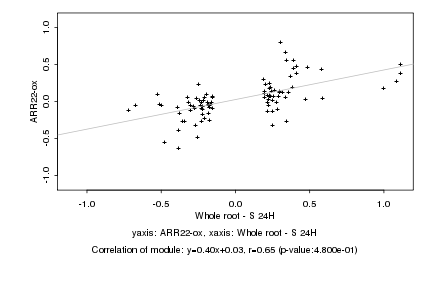
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Whole root - S 24H |
Log2 signal ratio
ARR22-ox |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.109 |
0.384 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.106 |
0.503 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
1.082 |
0.275 |
| 4 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.997 |
0.187 |
| 5 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.581 |
0.050 |
| 6 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.573 |
0.437 |
| 7 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.485 |
0.472 |
| 8 |
255075_at |
AT4G09110
|
[RING/U-box superfamily protein] |
0.469 |
0.037 |
| 9 |
253525_at |
AT4G31330
|
unknown |
0.407 |
0.482 |
| 10 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.406 |
0.390 |
| 11 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.390 |
0.452 |
| 12 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.389 |
0.556 |
| 13 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.379 |
0.201 |
| 14 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.367 |
0.341 |
| 15 |
256454_at |
AT1G75280
|
[NmrA-like negative transcriptional regulator family protein] |
0.353 |
0.123 |
| 16 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.341 |
0.565 |
| 17 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.340 |
-0.262 |
| 18 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.334 |
0.667 |
| 19 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.333 |
0.056 |
| 20 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.315 |
0.126 |
| 21 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.297 |
0.807 |
| 22 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.295 |
0.147 |
| 23 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.295 |
0.135 |
| 24 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.287 |
0.073 |
| 25 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.279 |
-0.096 |
| 26 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
0.273 |
-0.001 |
| 27 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.258 |
0.158 |
| 28 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.256 |
0.069 |
| 29 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.244 |
-0.320 |
| 30 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.244 |
-0.123 |
| 31 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.243 |
0.020 |
| 32 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.237 |
0.136 |
| 33 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.234 |
0.196 |
| 34 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.230 |
0.070 |
| 35 |
263892_at |
AT2G36890
|
ATMYB38, MYB DOMAIN PROTEIN 38, BIT1, BLUE INSENSITIVE TRAIT 1, MYB38, MYB DOMAIN PROTEIN 38, RAX2, REGULATOR OF AXILLARY MERISTEMS 2 |
0.229 |
0.073 |
| 36 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
0.227 |
0.086 |
| 37 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.223 |
0.256 |
| 38 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.223 |
0.182 |
| 39 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.221 |
-0.043 |
| 40 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.221 |
0.029 |
| 41 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.215 |
-0.011 |
| 42 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.214 |
-0.123 |
| 43 |
246584_at |
AT5G14730
|
unknown |
0.214 |
0.091 |
| 44 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.200 |
0.243 |
| 45 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.192 |
0.118 |
| 46 |
248481_at |
AT5G50930
|
[Histone superfamily protein] |
0.192 |
0.057 |
| 47 |
256453_at |
AT1G75270
|
DHAR2, dehydroascorbate reductase 2 |
0.191 |
0.145 |
| 48 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.187 |
0.298 |
| 49 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.721 |
-0.112 |
| 50 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.674 |
-0.042 |
| 51 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.528 |
0.106 |
| 52 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.518 |
-0.031 |
| 53 |
265611_at |
AT2G25510
|
unknown |
-0.502 |
-0.048 |
| 54 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.482 |
-0.543 |
| 55 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.395 |
-0.071 |
| 56 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.390 |
-0.381 |
| 57 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.385 |
-0.632 |
| 58 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.380 |
-0.160 |
| 59 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.359 |
-0.259 |
| 60 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.344 |
-0.261 |
| 61 |
263737_at |
AT1G60010
|
unknown |
-0.323 |
0.065 |
| 62 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.319 |
-0.004 |
| 63 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.304 |
-0.048 |
| 64 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.303 |
-0.113 |
| 65 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.283 |
-0.064 |
| 66 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.279 |
-0.082 |
| 67 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.275 |
-0.322 |
| 68 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
-0.263 |
0.043 |
| 69 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.258 |
-0.480 |
| 70 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.250 |
0.241 |
| 71 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.244 |
0.016 |
| 72 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.239 |
-0.045 |
| 73 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.235 |
-0.008 |
| 74 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
-0.232 |
-0.083 |
| 75 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.231 |
-0.259 |
| 76 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.223 |
-0.101 |
| 77 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.222 |
-0.054 |
| 78 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.222 |
-0.175 |
| 79 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
-0.218 |
0.014 |
| 80 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.213 |
0.058 |
| 81 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.212 |
-0.222 |
| 82 |
248186_at |
AT5G53880
|
unknown |
-0.198 |
0.103 |
| 83 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
-0.191 |
-0.010 |
| 84 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.186 |
-0.037 |
| 85 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.185 |
-0.159 |
| 86 |
252987_at |
AT4G38390
|
RHS17, root hair specific 17 |
-0.182 |
-0.048 |
| 87 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-0.178 |
-0.070 |
| 88 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.177 |
-0.044 |
| 89 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.176 |
-0.249 |
| 90 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.175 |
-0.076 |
| 91 |
264781_at |
AT1G08540
|
ABC1, ATSIG1, SIGMA FACTOR 1, ATSIG2, SIGMA FACTOR 2, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, SIGB, SIGMA FACTOR B |
-0.173 |
-0.028 |
| 92 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
-0.167 |
-0.010 |
| 93 |
250685_at |
AT5G06670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.161 |
0.057 |
| 94 |
251310_at |
AT3G61150
|
HD-GL2-1, HOMEODOMAIN-GLABRA2 1, HDG1, homeodomain GLABROUS 1 |
-0.161 |
0.081 |
| 95 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.159 |
-0.093 |