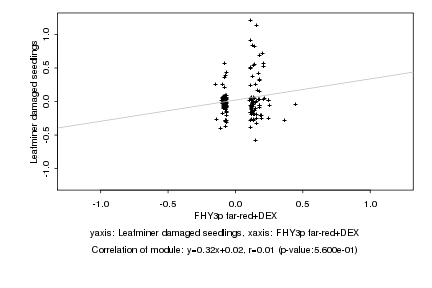
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
FHY3p far-red+DEX |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
267580_at |
AT2G41990
|
unknown |
0.439 |
-0.044 |
| 2 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
0.362 |
-0.268 |
| 3 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
0.247 |
-0.054 |
| 4 |
251610_at |
AT3G57930
|
unknown |
0.245 |
0.017 |
| 5 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
0.244 |
-0.242 |
| 6 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.242 |
0.018 |
| 7 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
0.212 |
0.055 |
| 8 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.203 |
0.576 |
| 9 |
257413_at |
AT1G22910
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.203 |
0.035 |
| 10 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.201 |
0.528 |
| 11 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.199 |
0.716 |
| 12 |
251365_at |
AT3G61310
|
AHL11, AT-hook motif nuclear localized protein 11 |
0.192 |
-0.246 |
| 13 |
259733_at |
AT1G77480
|
[Eukaryotic aspartyl protease family protein] |
0.187 |
-0.199 |
| 14 |
248764_at |
AT5G47640
|
NF-YB2, nuclear factor Y, subunit B2 |
0.182 |
-0.207 |
| 15 |
251541_at |
AT3G58750
|
CSY2, citrate synthase 2 |
0.178 |
0.154 |
| 16 |
266809_at |
AT2G29970
|
SMXL7, SMAX1-like 7 |
0.177 |
-0.059 |
| 17 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
0.175 |
-0.080 |
| 18 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.174 |
0.328 |
| 19 |
245617_at |
AT4G14490
|
[SMAD/FHA domain-containing protein ] |
0.174 |
-0.055 |
| 20 |
256884_at |
AT3G15200
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.172 |
0.331 |
| 21 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.172 |
0.697 |
| 22 |
261101_at |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
0.172 |
0.038 |
| 23 |
262550_at |
AT1G31310
|
[hydroxyproline-rich glycoprotein family protein] |
0.171 |
0.037 |
| 24 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.168 |
0.422 |
| 25 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.162 |
0.172 |
| 26 |
250327_at |
AT5G12050
|
unknown |
0.156 |
-0.252 |
| 27 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
0.155 |
-0.327 |
| 28 |
259050_at |
AT3G03360
|
[F-box/RNI-like superfamily protein] |
0.153 |
-0.191 |
| 29 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.152 |
1.146 |
| 30 |
247464_at |
AT5G62070
|
IQD23, IQ-domain 23 |
0.152 |
0.012 |
| 31 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
0.147 |
-0.099 |
| 32 |
250145_at |
AT5G14690
|
unknown |
0.147 |
-0.008 |
| 33 |
262811_at |
AT1G11700
|
unknown |
0.146 |
-0.572 |
| 34 |
258078_at |
AT3G25870
|
unknown |
0.142 |
0.257 |
| 35 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.139 |
0.556 |
| 36 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.139 |
0.053 |
| 37 |
251057_at |
AT5G01780
|
[2-oxoglutarate-dependent dioxygenase family protein] |
0.139 |
-0.131 |
| 38 |
257427_at |
AT1G79060
|
unknown |
0.138 |
-0.185 |
| 39 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.135 |
0.829 |
| 40 |
246911_at |
AT5G25810
|
tny, TINY |
0.134 |
-0.000 |
| 41 |
259755_at |
AT1G71070
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.133 |
-0.031 |
| 42 |
247349_at |
AT5G63820
|
unknown |
0.132 |
0.064 |
| 43 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.132 |
0.543 |
| 44 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.131 |
-0.265 |
| 45 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
0.131 |
-0.171 |
| 46 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.129 |
-0.268 |
| 47 |
245382_at |
AT4G17800
|
AHL23, AT-hook motif nuclear localized protein 23 |
0.128 |
0.015 |
| 48 |
254362_at |
AT4G22160
|
unknown |
0.127 |
-0.180 |
| 49 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.126 |
0.381 |
| 50 |
266880_at |
AT2G44770
|
[ELMO/CED-12 family protein] |
0.125 |
-0.022 |
| 51 |
250106_at |
AT5G16640
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.125 |
-0.143 |
| 52 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.124 |
0.843 |
| 53 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.122 |
-0.062 |
| 54 |
251326_at |
AT3G61590
|
HS, HWS, HAWAIIAN SKIRT |
0.121 |
0.000 |
| 55 |
245269_at |
AT4G14500
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.119 |
0.039 |
| 56 |
256282_at |
AT3G12550
|
FDM3, factor of DNA methylation 3 |
0.119 |
-0.084 |
| 57 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
0.119 |
-0.191 |
| 58 |
262512_at |
AT1G17145
|
[RING/U-box superfamily protein] |
0.119 |
0.071 |
| 59 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
0.117 |
-0.256 |
| 60 |
267561_at |
AT2G45590
|
[Protein kinase superfamily protein] |
0.117 |
-0.112 |
| 61 |
251206_at |
AT3G63090
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.116 |
0.025 |
| 62 |
267077_at |
AT2G40970
|
MYBC1 |
0.116 |
-0.103 |
| 63 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.116 |
-0.062 |
| 64 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
0.112 |
-0.070 |
| 65 |
265661_at |
AT2G24360
|
[Protein kinase superfamily protein] |
0.111 |
0.049 |
| 66 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.111 |
0.500 |
| 67 |
260744_at |
AT1G15010
|
unknown |
0.109 |
0.917 |
| 68 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.109 |
0.514 |
| 69 |
246796_at |
AT5G26770
|
unknown |
0.109 |
0.249 |
| 70 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.108 |
-0.152 |
| 71 |
261853_at |
AT1G50660
|
unknown |
0.108 |
-0.162 |
| 72 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.107 |
-0.373 |
| 73 |
255424_at |
AT4G03340
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.106 |
-0.054 |
| 74 |
250901_at |
AT5G03530
|
ATRAB, ATRAB ALPHA, ATRAB18B, ATRABC2A, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG C2A, RABC2A, RAB GTPase homolog C2A |
0.105 |
-0.274 |
| 75 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.105 |
1.222 |
| 76 |
245742_at |
AT1G44170
|
ALDH3H1, aldehyde dehydrogenase 3H1, ALDH4, aldehyde dehydrogenase 4 |
0.102 |
0.019 |
| 77 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.153 |
0.265 |
| 78 |
265308_at |
AT2G20300
|
ALE2, Abnormal Leaf Shape 2 |
-0.144 |
-0.259 |
| 79 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.116 |
-0.398 |
| 80 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
-0.107 |
0.020 |
| 81 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
-0.103 |
0.265 |
| 82 |
247231_at |
AT5G65230
|
AtMYB53, myb domain protein 53, MYB53, myb domain protein 53 |
-0.101 |
-0.026 |
| 83 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.100 |
-0.166 |
| 84 |
253713_at |
AT4G29370
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.100 |
-0.013 |
| 85 |
251925_at |
AT3G54000
|
unknown |
-0.099 |
0.060 |
| 86 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.097 |
0.058 |
| 87 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.094 |
-0.054 |
| 88 |
262069_at |
AT1G80090
|
CBSX4, CBS domain containing protein 4 |
-0.094 |
-0.062 |
| 89 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-0.094 |
-0.075 |
| 90 |
254193_at |
AT4G23870
|
unknown |
-0.094 |
0.036 |
| 91 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
-0.092 |
-0.031 |
| 92 |
247070_at |
AT5G66815
|
unknown |
-0.092 |
0.005 |
| 93 |
256926_at |
AT3G22540
|
unknown |
-0.091 |
-0.100 |
| 94 |
258685_at |
AT3G07830
|
PGA3, polygalacturonase 3 |
-0.091 |
0.046 |
| 95 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.090 |
0.088 |
| 96 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
-0.088 |
0.089 |
| 97 |
257843_at |
AT3G28400
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 4.6e-39 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
-0.088 |
-0.003 |
| 98 |
261641_at |
AT1G27670
|
unknown |
-0.088 |
-0.028 |
| 99 |
266739_at |
AT2G46730
|
[pseudogene] |
-0.087 |
-0.078 |
| 100 |
254646_at |
AT4G18530
|
unknown |
-0.086 |
0.369 |
| 101 |
263528_at |
AT2G24800
|
[Peroxidase superfamily protein] |
-0.086 |
0.017 |
| 102 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.085 |
0.215 |
| 103 |
253192_at |
AT4G35370
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.085 |
-0.052 |
| 104 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.085 |
-0.028 |
| 105 |
259982_at |
AT1G76410
|
ATL8 |
-0.084 |
0.077 |
| 106 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
-0.084 |
-0.039 |
| 107 |
257231_at |
AT3G16550
|
DEG12, degradation of periplasmic proteins 12, DEGP12, DEGP protease 12 |
-0.083 |
0.015 |
| 108 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.082 |
0.569 |
| 109 |
261035_at |
AT1G17390
|
[similar to reverse transcriptase-related [Arabidopsis thaliana] (TAIR:AT4G12275.1)] |
-0.082 |
0.029 |
| 110 |
258959_at |
AT3G10600
|
CAT7, cationic amino acid transporter 7 |
-0.082 |
-0.040 |
| 111 |
266543_at |
AT2G35075
|
unknown |
-0.081 |
0.030 |
| 112 |
260631_at |
AT1G62350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.081 |
0.000 |
| 113 |
263872_at |
AT2G21770
|
CESA09, CESA9, cellulose synthase A9 |
-0.080 |
-0.044 |
| 114 |
246626_at |
AT1G48870
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.079 |
-0.054 |
| 115 |
260935_at |
AT1G45110
|
[Tetrapyrrole (Corrin/Porphyrin) Methylases] |
-0.079 |
0.007 |
| 116 |
262872_at |
AT1G64690
|
BLT, BRANCHLESS TRICHOMES |
-0.078 |
-0.278 |
| 117 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.078 |
-0.104 |
| 118 |
245254_at |
AT4G14680
|
APS3 |
-0.078 |
-0.010 |
| 119 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.078 |
0.395 |
| 120 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.078 |
-0.094 |
| 121 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.077 |
-0.287 |
| 122 |
263176_at |
AT1G05530
|
UGT2, UDP-GLUCOSYL TRANSFERASE 2, UGT75B2, UDP-glucosyl transferase 75B2 |
-0.076 |
-0.042 |
| 123 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.076 |
-0.270 |
| 124 |
263721_at |
AT2G13630
|
[F-box associated ubiquitination effector family protein] |
-0.076 |
0.034 |
| 125 |
256420_at |
AT1G33475
|
[SNARE-like superfamily protein] |
-0.076 |
-0.085 |
| 126 |
258266_at |
AT3G15860
|
unknown |
-0.076 |
-0.011 |
| 127 |
258543_at |
AT3G06870
|
[proline-rich family protein] |
-0.075 |
-0.036 |
| 128 |
248292_at |
AT5G53030
|
unknown |
-0.075 |
0.101 |
| 129 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.075 |
-0.360 |
| 130 |
263730_at |
AT1G60090
|
BGLU4, beta glucosidase 4 |
-0.075 |
0.001 |
| 131 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.074 |
0.002 |
| 132 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.074 |
-0.112 |
| 133 |
246552_at |
AT5G15420
|
unknown |
-0.074 |
-0.020 |
| 134 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.074 |
-0.024 |
| 135 |
264813_at |
AT2G17890
|
CPK16, calcium-dependent protein kinase 16 |
-0.074 |
-0.048 |
| 136 |
267281_at |
AT2G19400
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.072 |
-0.073 |
| 137 |
250136_at |
AT5G15380
|
DRM1, domains rearranged methylase 1 |
-0.072 |
-0.020 |
| 138 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.072 |
-0.053 |
| 139 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.071 |
0.048 |
| 140 |
262918_at |
AT1G65000
|
unknown |
-0.071 |
0.097 |
| 141 |
260074_at |
AT1G73640
|
AtRABA6a, RAB GTPase homolog A6A, RABA6a, RAB GTPase homolog A6A |
-0.071 |
-0.065 |
| 142 |
251330_at |
AT3G61550
|
[RING/U-box superfamily protein] |
-0.070 |
-0.082 |
| 143 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.070 |
-0.279 |
| 144 |
259220_at |
AT3G03550
|
[RING/U-box superfamily protein] |
-0.070 |
0.042 |
| 145 |
262315_at |
AT1G70990
|
[proline-rich family protein] |
-0.070 |
-0.004 |
| 146 |
249211_at |
AT5G42680
|
unknown |
-0.069 |
-0.300 |
| 147 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
-0.069 |
0.443 |
| 148 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.069 |
-0.198 |
| 149 |
248265_at |
AT5G53430
|
ATX5, SDG29, SET domain group 29, SET29 |
-0.068 |
-0.160 |
| 150 |
245824_at |
AT1G57943
|
ATPUP17, purine permease 17, PUP17, purine permease 17 |
-0.068 |
0.067 |
| 151 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.067 |
-0.145 |