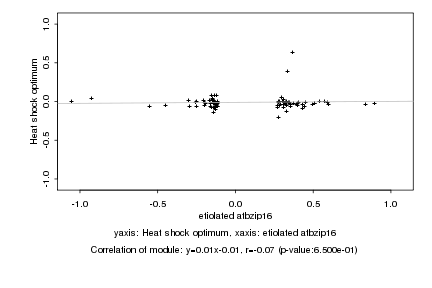
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
etiolated atbzip16 |
Log2 signal ratio
Heat shock optimum |
| 1 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
0.888 |
-0.024 |
| 2 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
0.834 |
-0.029 |
| 3 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.593 |
-0.029 |
| 4 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
0.592 |
-0.007 |
| 5 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
0.572 |
0.011 |
| 6 |
246925_at |
AT5G25180
|
CYP71B14, cytochrome P450, family 71, subfamily B, polypeptide 14 |
0.537 |
0.010 |
| 7 |
255414_at |
AT4G03156
|
[small GTPase-related] |
0.507 |
-0.016 |
| 8 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.506 |
-0.020 |
| 9 |
267321_at |
AT2G19320
|
unknown |
0.495 |
-0.030 |
| 10 |
256139_at |
AT1G48660
|
[Auxin-responsive GH3 family protein] |
0.445 |
-0.004 |
| 11 |
255840_at |
AT2G33520
|
unknown |
0.444 |
-0.055 |
| 12 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
0.425 |
-0.086 |
| 13 |
262010_at |
AT1G35612
|
[expressed protein] |
0.425 |
-0.034 |
| 14 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
0.401 |
-0.012 |
| 15 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
0.396 |
-0.016 |
| 16 |
251202_at |
AT3G63040
|
unknown |
0.394 |
-0.043 |
| 17 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
0.388 |
-0.035 |
| 18 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.368 |
-0.015 |
| 19 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.362 |
0.640 |
| 20 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
0.352 |
-0.027 |
| 21 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.352 |
-0.031 |
| 22 |
265714_at |
AT2G03520
|
ATUPS4, ureide permease 4, UPS4, ureide permease 4 |
0.352 |
-0.064 |
| 23 |
247226_at |
AT5G65100
|
[Ethylene insensitive 3 family protein] |
0.343 |
-0.007 |
| 24 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
0.335 |
-0.009 |
| 25 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.330 |
0.394 |
| 26 |
251350_at |
AT3G61040
|
CYP76C7, cytochrome P450, family 76, subfamily C, polypeptide 7 |
0.327 |
-0.044 |
| 27 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
0.327 |
0.006 |
| 28 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
0.325 |
-0.123 |
| 29 |
265123_at |
AT1G55440
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.323 |
-0.006 |
| 30 |
247172_at |
AT5G65550
|
[UDP-Glycosyltransferase superfamily protein] |
0.321 |
-0.030 |
| 31 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.320 |
-0.012 |
| 32 |
253870_at |
AT4G27530
|
unknown |
0.317 |
-0.022 |
| 33 |
245161_at |
AT2G33070
|
ATNSP2, NITRILE-SPECIFIER PROTEIN 2, NSP2, nitrile specifier protein 2 |
0.308 |
0.035 |
| 34 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.307 |
-0.027 |
| 35 |
248096_at |
AT5G55240
|
ATPXG2, ARABIDOPSIS THALIANA PEROXYGENASE 2 |
0.305 |
0.002 |
| 36 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.303 |
-0.065 |
| 37 |
255049_at |
AT4G09610
|
GASA2, GAST1 protein homolog 2 |
0.299 |
0.005 |
| 38 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.297 |
0.006 |
| 39 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.295 |
0.058 |
| 40 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.285 |
-0.029 |
| 41 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.282 |
0.009 |
| 42 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.279 |
-0.039 |
| 43 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.273 |
-0.195 |
| 44 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.272 |
0.008 |
| 45 |
266540_at |
AT2G35310
|
[Transcriptional factor B3 family protein] |
0.271 |
-0.003 |
| 46 |
248210_at |
AT5G54000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.270 |
-0.051 |
| 47 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
0.268 |
-0.071 |
| 48 |
260835_at |
AT1G06700
|
[Protein kinase superfamily protein] |
-1.059 |
0.001 |
| 49 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.927 |
0.040 |
| 50 |
262634_at |
AT1G06690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.553 |
-0.058 |
| 51 |
266634_at |
AT2G35530
|
AtbZIP16, bZIP16, basic region/leucine zipper transcription factor 16 |
-0.455 |
-0.050 |
| 52 |
259464_at |
AT1G18990
|
unknown |
-0.303 |
0.015 |
| 53 |
262636_at |
AT1G06670
|
NIH, nuclear DEIH-boxhelicase |
-0.297 |
-0.060 |
| 54 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
-0.263 |
0.000 |
| 55 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.255 |
-0.056 |
| 56 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.254 |
0.003 |
| 57 |
251372_at |
AT3G60520
|
unknown |
-0.211 |
0.016 |
| 58 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.203 |
-0.045 |
| 59 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.201 |
-0.008 |
| 60 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
-0.195 |
-0.020 |
| 61 |
261345_at |
AT1G79760
|
DTA4, downstream target of AGL15-4 |
-0.168 |
0.023 |
| 62 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.165 |
-0.058 |
| 63 |
255401_at |
AT4G03600
|
unknown |
-0.163 |
-0.024 |
| 64 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.160 |
0.083 |
| 65 |
254876_at |
AT4G11610
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.159 |
-0.065 |
| 66 |
248863_at |
AT5G46750
|
AGD9, ARF-GAP domain 9 |
-0.151 |
-0.000 |
| 67 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.150 |
0.047 |
| 68 |
248864_at |
AT5G46760
|
MYC3 |
-0.150 |
0.016 |
| 69 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
-0.148 |
0.037 |
| 70 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.147 |
0.024 |
| 71 |
260583_x_at |
AT2G47220
|
unknown |
-0.145 |
-0.133 |
| 72 |
264896_at |
AT1G23210
|
AtGH9B6, glycosyl hydrolase 9B6, GH9B6, glycosyl hydrolase 9B6 |
-0.143 |
-0.014 |
| 73 |
262974_at |
AT1G75550
|
[glycine-rich protein] |
-0.143 |
-0.073 |
| 74 |
248670_at |
AT5G48740
|
[Leucine-rich repeat protein kinase family protein] |
-0.142 |
0.021 |
| 75 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.141 |
0.087 |
| 76 |
266673_at |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
-0.140 |
0.010 |
| 77 |
258095_at |
AT3G23610
|
DSPTP1, dual specificity protein phosphatase 1 |
-0.136 |
-0.068 |
| 78 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.135 |
-0.015 |
| 79 |
246883_at |
AT5G26190
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.134 |
-0.099 |
| 80 |
266710_at |
AT2G46850
|
[Protein kinase superfamily protein] |
-0.132 |
-0.038 |
| 81 |
261096_at |
AT1G62940
|
ACOS5, acyl-CoA synthetase 5 |
-0.129 |
-0.055 |
| 82 |
254412_at |
AT4G21430
|
B160 |
-0.128 |
-0.017 |
| 83 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.126 |
-0.027 |
| 84 |
262965_at |
AT1G54310
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.125 |
-0.015 |
| 85 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
-0.124 |
-0.061 |
| 86 |
263712_at |
AT2G20585
|
NFD6, NUCLEAR FUSION DEFECTIVE 6 |
-0.124 |
0.080 |
| 87 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.124 |
-0.049 |
| 88 |
266262_at |
AT2G27590
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.122 |
-0.035 |
| 89 |
252181_at |
AT3G50685
|
unknown |
-0.121 |
-0.022 |
| 90 |
247970_at |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
-0.121 |
-0.059 |
| 91 |
249509_at |
AT5G38390
|
[F-box/RNI-like superfamily protein] |
-0.120 |
-0.054 |
| 92 |
247027_at |
AT5G67090
|
[Subtilisin-like serine endopeptidase family protein] |
-0.118 |
-0.001 |
| 93 |
264104_at |
AT2G13750
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.5e-17 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.118 |
0.005 |
| 94 |
246911_at |
AT5G25810
|
tny, TINY |
-0.117 |
-0.010 |