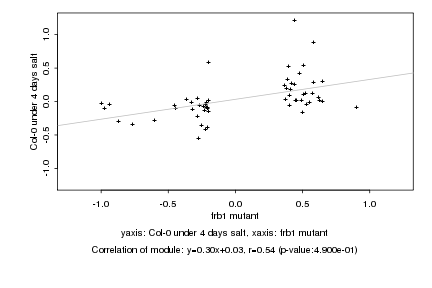
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
frb1 mutant |
Log2 signal ratio
Col-0 under 4 days salt |
| 1 |
257969_at |
AT3G27520
|
unknown |
0.900 |
-0.081 |
| 2 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.647 |
0.307 |
| 3 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.644 |
0.011 |
| 4 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.620 |
0.020 |
| 5 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.612 |
0.062 |
| 6 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.574 |
0.299 |
| 7 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.574 |
0.885 |
| 8 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.571 |
0.123 |
| 9 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
0.549 |
-0.006 |
| 10 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.523 |
-0.043 |
| 11 |
258327_at |
AT3G22640
|
PAP85 |
0.518 |
0.123 |
| 12 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
0.505 |
0.117 |
| 13 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.504 |
0.545 |
| 14 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.495 |
-0.158 |
| 15 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.487 |
0.022 |
| 16 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.476 |
0.425 |
| 17 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
0.454 |
0.019 |
| 18 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.446 |
0.027 |
| 19 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.439 |
0.262 |
| 20 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.436 |
1.226 |
| 21 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.412 |
0.283 |
| 22 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.409 |
0.188 |
| 23 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.402 |
0.100 |
| 24 |
247552_at |
AT5G60920
|
COB, COBRA |
0.396 |
-0.055 |
| 25 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.389 |
0.528 |
| 26 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.387 |
0.330 |
| 27 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.374 |
0.207 |
| 28 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.369 |
0.041 |
| 29 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.365 |
0.248 |
| 30 |
251122_at |
AT5G01020
|
[Protein kinase superfamily protein] |
-1.002 |
-0.025 |
| 31 |
251140_at |
AT5G01090
|
[Concanavalin A-like lectin family protein] |
-0.981 |
-0.092 |
| 32 |
245862_at |
AT5G01010
|
unknown |
-0.939 |
-0.033 |
| 33 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.878 |
-0.290 |
| 34 |
251142_at |
AT5G01015
|
unknown |
-0.773 |
-0.339 |
| 35 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.607 |
-0.276 |
| 36 |
250152_at |
AT5G15120
|
unknown |
-0.460 |
-0.050 |
| 37 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.448 |
-0.096 |
| 38 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.365 |
0.044 |
| 39 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.331 |
-0.004 |
| 40 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.325 |
-0.110 |
| 41 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.285 |
0.047 |
| 42 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
-0.284 |
-0.215 |
| 43 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.281 |
-0.544 |
| 44 |
254043_at |
AT4G25990
|
CIL |
-0.268 |
-0.050 |
| 45 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.260 |
-0.358 |
| 46 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.244 |
-0.065 |
| 47 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.234 |
-0.124 |
| 48 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.230 |
-0.404 |
| 49 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.225 |
-0.030 |
| 50 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.217 |
-0.079 |
| 51 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
-0.216 |
-0.007 |
| 52 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.215 |
-0.075 |
| 53 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.212 |
-0.090 |
| 54 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.209 |
-0.377 |
| 55 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.208 |
0.026 |
| 56 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.208 |
-0.145 |
| 57 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.205 |
0.590 |