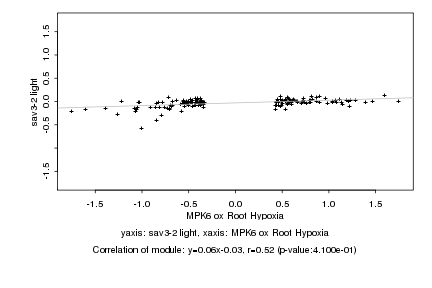
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
MPK6 ox Root Hypoxia |
Log2 signal ratio
sav3-2 light |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.742 |
0.005 |
| 2 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.586 |
0.139 |
| 3 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.458 |
0.001 |
| 4 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.381 |
-0.003 |
| 5 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.283 |
0.025 |
| 6 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.231 |
0.027 |
| 7 |
245226_at |
AT3G29970
|
[B12D protein] |
1.214 |
-0.092 |
| 8 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.208 |
0.007 |
| 9 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.205 |
0.012 |
| 10 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.183 |
0.023 |
| 11 |
257925_at |
AT3G23170
|
unknown |
1.143 |
-0.056 |
| 12 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.125 |
-0.001 |
| 13 |
254200_at |
AT4G24110
|
unknown |
1.104 |
0.050 |
| 14 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
1.080 |
-0.005 |
| 15 |
253616_at |
AT4G30380
|
[Barwin-related endoglucanase] |
1.060 |
0.031 |
| 16 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
1.035 |
-0.017 |
| 17 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
1.033 |
0.004 |
| 18 |
249384_at |
AT5G39890
|
unknown |
0.985 |
-0.033 |
| 19 |
250152_at |
AT5G15120
|
unknown |
0.957 |
0.074 |
| 20 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.891 |
-0.007 |
| 21 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.889 |
0.127 |
| 22 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.865 |
0.014 |
| 23 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.859 |
0.097 |
| 24 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.814 |
0.051 |
| 25 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.806 |
0.121 |
| 26 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.800 |
0.034 |
| 27 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.796 |
-0.016 |
| 28 |
266998_at |
AT2G34400
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.790 |
-0.001 |
| 29 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.783 |
0.042 |
| 30 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.759 |
-0.041 |
| 31 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.731 |
-0.004 |
| 32 |
246200_at |
AT4G37240
|
unknown |
0.723 |
0.031 |
| 33 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.718 |
0.076 |
| 34 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.716 |
-0.016 |
| 35 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.713 |
0.003 |
| 36 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.706 |
-0.035 |
| 37 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.664 |
0.021 |
| 38 |
266875_at |
AT2G44800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.657 |
-0.015 |
| 39 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.644 |
0.021 |
| 40 |
258991_at |
AT3G08820
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.622 |
0.034 |
| 41 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.617 |
0.057 |
| 42 |
255532_at |
AT4G02170
|
unknown |
0.602 |
0.042 |
| 43 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.598 |
-0.043 |
| 44 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.592 |
0.006 |
| 45 |
245401_at |
AT4G17670
|
unknown |
0.590 |
-0.003 |
| 46 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.586 |
0.012 |
| 47 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.585 |
0.018 |
| 48 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.580 |
0.009 |
| 49 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.575 |
0.021 |
| 50 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.574 |
0.064 |
| 51 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.573 |
0.020 |
| 52 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
0.572 |
0.017 |
| 53 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.567 |
-0.014 |
| 54 |
256262_at |
AT3G12150
|
unknown |
0.565 |
-0.034 |
| 55 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.565 |
0.068 |
| 56 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
0.555 |
-0.049 |
| 57 |
262392_at |
AT1G49520
|
[SWIB complex BAF60b domain-containing protein] |
0.554 |
0.093 |
| 58 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.552 |
-0.037 |
| 59 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.535 |
0.025 |
| 60 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.535 |
-0.150 |
| 61 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.535 |
0.062 |
| 62 |
247024_at |
AT5G66985
|
unknown |
0.532 |
0.014 |
| 63 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.500 |
0.009 |
| 64 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
0.498 |
-0.036 |
| 65 |
259996_at |
AT1G67910
|
unknown |
0.496 |
0.026 |
| 66 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.492 |
0.015 |
| 67 |
257466_at |
AT1G62840
|
unknown |
0.490 |
0.005 |
| 68 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.484 |
-0.080 |
| 69 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.481 |
0.060 |
| 70 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
0.480 |
-0.097 |
| 71 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.473 |
0.111 |
| 72 |
260005_at |
AT1G67920
|
unknown |
0.458 |
-0.016 |
| 73 |
266220_at |
AT2G28755
|
[UDP-D-glucuronate carboxy-lyase-related] |
0.454 |
0.004 |
| 74 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.451 |
-0.065 |
| 75 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.450 |
0.045 |
| 76 |
246158_at |
AT5G19855
|
AtRbcX2, RbcX2, homologue of cyanobacterial RbcX 2 |
0.446 |
0.019 |
| 77 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.440 |
0.003 |
| 78 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.438 |
-0.007 |
| 79 |
264836_at |
AT1G03610
|
unknown |
0.434 |
-0.079 |
| 80 |
258591_at |
AT3G04360
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.434 |
-0.018 |
| 81 |
248820_at |
AT5G47060
|
unknown |
0.431 |
0.003 |
| 82 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.428 |
-0.155 |
| 83 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.428 |
-0.064 |
| 84 |
266393_at |
AT2G41260
|
ATM17, M17 |
-1.762 |
-0.198 |
| 85 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-1.611 |
-0.169 |
| 86 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
-1.395 |
-0.146 |
| 87 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-1.269 |
-0.274 |
| 88 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
-1.220 |
0.001 |
| 89 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
-1.081 |
-0.138 |
| 90 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-1.070 |
-0.196 |
| 91 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-1.068 |
-0.170 |
| 92 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-1.049 |
-0.116 |
| 93 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-1.046 |
-0.003 |
| 94 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
-1.037 |
-0.016 |
| 95 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-1.014 |
-0.567 |
| 96 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.912 |
-0.109 |
| 97 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.857 |
-0.126 |
| 98 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.853 |
-0.387 |
| 99 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.848 |
-0.034 |
| 100 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.826 |
-0.015 |
| 101 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
-0.822 |
-0.114 |
| 102 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.796 |
-0.296 |
| 103 |
245161_at |
AT2G33070
|
ATNSP2, NITRILE-SPECIFIER PROTEIN 2, NSP2, nitrile specifier protein 2 |
-0.784 |
-0.019 |
| 104 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.766 |
-0.112 |
| 105 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.735 |
-0.133 |
| 106 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.726 |
0.101 |
| 107 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.711 |
-0.156 |
| 108 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.708 |
-0.133 |
| 109 |
258327_at |
AT3G22640
|
PAP85 |
-0.698 |
-0.078 |
| 110 |
259615_at |
AT1G47980
|
unknown |
-0.685 |
-0.087 |
| 111 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.681 |
-0.081 |
| 112 |
258489_at |
AT3G02520
|
GF14 NU, GRF7, general regulatory factor 7 |
-0.678 |
0.005 |
| 113 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
-0.640 |
0.040 |
| 114 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
-0.588 |
-0.061 |
| 115 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.586 |
-0.205 |
| 116 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
-0.572 |
-0.023 |
| 117 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.563 |
0.005 |
| 118 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.561 |
0.040 |
| 119 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.552 |
-0.062 |
| 120 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.552 |
-0.001 |
| 121 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.547 |
-0.031 |
| 122 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.547 |
-0.087 |
| 123 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.534 |
-0.034 |
| 124 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.529 |
0.015 |
| 125 |
259578_at |
AT1G27990
|
unknown |
-0.529 |
-0.017 |
| 126 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.523 |
-0.033 |
| 127 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
-0.517 |
-0.055 |
| 128 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.515 |
-0.021 |
| 129 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.505 |
0.001 |
| 130 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.503 |
-0.067 |
| 131 |
255763_at |
AT1G16730
|
unknown |
-0.490 |
0.064 |
| 132 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.483 |
-0.009 |
| 133 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.478 |
-0.005 |
| 134 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.469 |
-0.104 |
| 135 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.466 |
0.032 |
| 136 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.461 |
-0.020 |
| 137 |
261247_at |
AT1G20070
|
unknown |
-0.443 |
-0.064 |
| 138 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.440 |
-0.076 |
| 139 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.436 |
-0.064 |
| 140 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.433 |
-0.008 |
| 141 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
-0.431 |
-0.073 |
| 142 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.428 |
0.067 |
| 143 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.426 |
0.022 |
| 144 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
-0.421 |
-0.008 |
| 145 |
253044_at |
AT4G37290
|
unknown |
-0.414 |
0.074 |
| 146 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.411 |
0.053 |
| 147 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.407 |
-0.013 |
| 148 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.402 |
-0.034 |
| 149 |
264537_at |
AT1G55610
|
BRL1, BRI1 like |
-0.399 |
-0.079 |
| 150 |
262396_at |
AT1G49470
|
unknown |
-0.399 |
-0.032 |
| 151 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
-0.389 |
-0.010 |
| 152 |
266222_at |
AT2G28780
|
unknown |
-0.381 |
-0.084 |
| 153 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.379 |
-0.010 |
| 154 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.377 |
0.069 |
| 155 |
266282_at |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
-0.377 |
-0.039 |
| 156 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
-0.376 |
-0.060 |
| 157 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.375 |
-0.014 |
| 158 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.371 |
0.010 |
| 159 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.368 |
0.028 |
| 160 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.366 |
0.036 |
| 161 |
245082_at |
AT2G23270
|
unknown |
-0.354 |
-0.040 |
| 162 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.352 |
-0.118 |
| 163 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.350 |
-0.043 |
| 164 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.343 |
-0.002 |
| 165 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.333 |
-0.009 |