|
probeID |
AGICode |
Annotation |
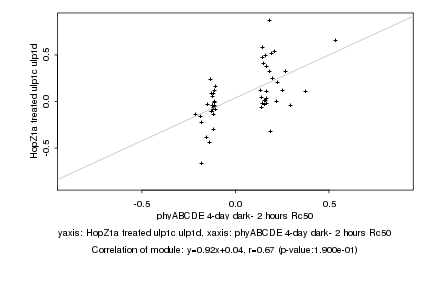
Log2 signal ratio
phyABCDE 4-day dark- 2 hours Rc50 |
Log2 signal ratio
HopZ1a treated ulp1c ulp1d |
| 1 |
248218_at |
AT5G53710
|
unknown |
0.532 |
0.661 |
| 2 |
251377_at |
AT3G60650
|
unknown |
0.374 |
0.108 |
| 3 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.292 |
-0.038 |
| 4 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.264 |
0.324 |
| 5 |
263285_at |
AT2G36120
|
DOT1, DEFECTIVELY ORGANIZED TRIBUTARIES 1 |
0.250 |
0.126 |
| 6 |
259985_at |
AT1G76620
|
unknown |
0.220 |
0.207 |
| 7 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.215 |
0.004 |
| 8 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.204 |
0.540 |
| 9 |
256759_at |
AT3G25640
|
unknown |
0.193 |
0.255 |
| 10 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.190 |
0.520 |
| 11 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.184 |
-0.318 |
| 12 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.179 |
0.880 |
| 13 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.179 |
0.325 |
| 14 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.165 |
0.381 |
| 15 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.164 |
0.112 |
| 16 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
0.163 |
-0.019 |
| 17 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
0.161 |
0.037 |
| 18 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.160 |
0.502 |
| 19 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
0.156 |
0.021 |
| 20 |
256990_at |
AT3G28590
|
unknown |
0.153 |
0.015 |
| 21 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.152 |
-0.028 |
| 22 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.147 |
0.411 |
| 23 |
249357_at |
AT5G40490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.143 |
-0.014 |
| 24 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.143 |
0.474 |
| 25 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.142 |
0.585 |
| 26 |
252665_at |
AT3G44140
|
unknown |
0.134 |
0.052 |
| 27 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
0.134 |
-0.055 |
| 28 |
253131_at |
AT4G35550
|
ATWOX13, HB-4, WOX13, WUSCHEL related homeobox 13 |
0.131 |
0.127 |
| 29 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.218 |
-0.130 |
| 30 |
265609_at |
AT2G25420
|
[transducin family protein / WD-40 repeat family protein] |
-0.188 |
-0.153 |
| 31 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.184 |
-0.215 |
| 32 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.183 |
-0.663 |
| 33 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.156 |
-0.385 |
| 34 |
256912_at |
AT3G23850
|
unknown |
-0.150 |
-0.025 |
| 35 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.140 |
-0.437 |
| 36 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
-0.134 |
0.238 |
| 37 |
263104_at |
AT2G05120
|
[Nucleoporin, Nup133/Nup155-like] |
-0.131 |
-0.104 |
| 38 |
245869_at |
AT1G26330
|
[DNA binding] |
-0.129 |
-0.106 |
| 39 |
261786_at |
AT1G15990
|
ATCNGC7, cyclic nucleotide gated channel 7, CNGC7, cyclic nucleotide gated channel 7 |
-0.128 |
0.089 |
| 40 |
267347_at |
AT2G39950
|
unknown |
-0.127 |
-0.041 |
| 41 |
258910_at |
AT3G06480
|
[DEAD box RNA helicase family protein] |
-0.127 |
-0.042 |
| 42 |
250321_at |
AT5G12850
|
[CCCH-type zinc finger protein with ARM repeat domain] |
-0.126 |
-0.083 |
| 43 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.125 |
0.061 |
| 44 |
254879_at |
AT4G11670
|
unknown |
-0.120 |
0.087 |
| 45 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.120 |
-0.056 |
| 46 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
-0.119 |
-0.133 |
| 47 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.119 |
-0.290 |
| 48 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.117 |
-0.003 |
| 49 |
262492_at |
AT1G21630
|
[Calcium-binding EF hand family protein] |
-0.116 |
-0.043 |
| 50 |
261800_at |
AT1G30490
|
ATHB9, PHV, PHAVOLUTA |
-0.116 |
0.003 |
| 51 |
246897_at |
AT5G25560
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.115 |
-0.047 |
| 52 |
261945_at |
AT1G64530
|
NLP6, NIN-like protein 6 |
-0.113 |
-0.039 |
| 53 |
261584_at |
AT1G01040
|
ASU1, ABNORMAL SUSPENSOR 1, ATDCL1, DICER-LIKE 1, CAF, CARPEL FACTORY, DCL1, dicer-like 1, EMB60, EMBRYO DEFECTIVE 60, EMB76, EMBRYO DEFECTIVE 76, SIN1, SHORT INTEGUMENTS 1, SUS1, SUSPENSOR 1 |
-0.113 |
0.125 |
| 54 |
253762_at |
AT4G28830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.110 |
-0.084 |
| 55 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
-0.110 |
0.164 |