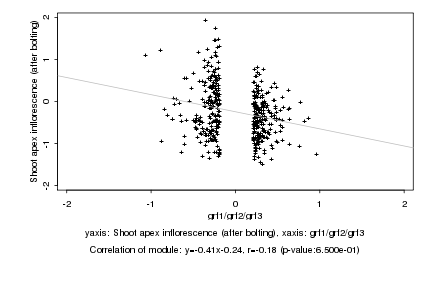
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
grf1/grf2/grf3 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
0.957 |
-1.243 |
| 2 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
0.865 |
-0.382 |
| 3 |
249687_at |
AT5G36150
|
ATPEN3, putative pentacyclic triterpene synthase 3, PEN3, putative pentacyclic triterpene synthase 3 |
0.811 |
-0.454 |
| 4 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.767 |
-0.009 |
| 5 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.751 |
-1.047 |
| 6 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
0.638 |
-0.150 |
| 7 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.635 |
-0.420 |
| 8 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.634 |
-1.003 |
| 9 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.622 |
-0.172 |
| 10 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.618 |
0.276 |
| 11 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.568 |
-0.128 |
| 12 |
256442_at |
AT3G10930
|
unknown |
0.553 |
-0.602 |
| 13 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
0.550 |
-0.925 |
| 14 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.548 |
-0.450 |
| 15 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.547 |
0.105 |
| 16 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.528 |
-0.561 |
| 17 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.528 |
-0.699 |
| 18 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.525 |
-0.210 |
| 19 |
262715_at |
AT1G16490
|
ATMYB58, MYB DOMAIN PROTEIN 58, MYB58, myb domain protein 58 |
0.514 |
-0.427 |
| 20 |
257490_x_at |
AT1G01380
|
ETC1, ENHANCER OF TRY AND CPC 1 |
0.493 |
-0.966 |
| 21 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.486 |
-0.432 |
| 22 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
0.485 |
0.338 |
| 23 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.484 |
-0.383 |
| 24 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.484 |
-0.246 |
| 25 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
0.477 |
-0.935 |
| 26 |
248011_at |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
0.470 |
-0.163 |
| 27 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.467 |
-0.722 |
| 28 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.465 |
-0.095 |
| 29 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.462 |
0.430 |
| 30 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.459 |
-0.390 |
| 31 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.458 |
-0.309 |
| 32 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.452 |
0.042 |
| 33 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.440 |
0.044 |
| 34 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.432 |
-1.111 |
| 35 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.426 |
0.336 |
| 36 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.424 |
-0.265 |
| 37 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.421 |
-1.379 |
| 38 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.420 |
-0.334 |
| 39 |
251466_at |
AT3G59340
|
unknown |
0.419 |
-0.524 |
| 40 |
264693_at |
AT1G69970
|
CLE26, CLAVATA3/ESR-RELATED 26 |
0.416 |
-1.038 |
| 41 |
263652_at |
AT1G04330
|
unknown |
0.404 |
-0.666 |
| 42 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.403 |
-0.249 |
| 43 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.401 |
-0.771 |
| 44 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.396 |
-0.525 |
| 45 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.388 |
-0.723 |
| 46 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.387 |
-0.203 |
| 47 |
253643_at |
AT4G29780
|
unknown |
0.383 |
0.234 |
| 48 |
267502_at |
AT2G45550
|
CYP76C4, cytochrome P450, family 76, subfamily C, polypeptide 4 |
0.379 |
-0.726 |
| 49 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.374 |
-0.863 |
| 50 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.370 |
-0.086 |
| 51 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.368 |
0.198 |
| 52 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.367 |
-0.138 |
| 53 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.361 |
-0.385 |
| 54 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.361 |
-0.855 |
| 55 |
251012_at |
AT5G02580
|
unknown |
0.352 |
-0.041 |
| 56 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.349 |
-0.429 |
| 57 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.347 |
-0.733 |
| 58 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.345 |
-0.439 |
| 59 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.344 |
-0.839 |
| 60 |
265062_at |
AT1G61550
|
[S-locus lectin protein kinase family protein] |
0.341 |
-1.143 |
| 61 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.340 |
-1.236 |
| 62 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.339 |
-0.417 |
| 63 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.337 |
0.014 |
| 64 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.335 |
-0.340 |
| 65 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
0.334 |
-0.684 |
| 66 |
258198_at |
AT3G14020
|
NF-YA6, nuclear factor Y, subunit A6 |
0.331 |
0.765 |
| 67 |
266900_at |
AT2G34610
|
unknown |
0.331 |
-0.197 |
| 68 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.330 |
-0.700 |
| 69 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.330 |
-0.091 |
| 70 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.329 |
-0.855 |
| 71 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.328 |
-0.114 |
| 72 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
0.325 |
-0.849 |
| 73 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.324 |
-0.136 |
| 74 |
265052_at |
AT1G51990
|
[O-methyltransferase family protein] |
0.323 |
-0.262 |
| 75 |
252163_at |
AT3G50610
|
unknown |
0.322 |
-0.544 |
| 76 |
245204_at |
AT5G12270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.321 |
-0.688 |
| 77 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.320 |
-0.064 |
| 78 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.319 |
-0.948 |
| 79 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.319 |
-0.006 |
| 80 |
246967_at |
AT5G24860
|
ATFPF1, ARABIDOPSIS FLOWERING PROMOTING FACTOR 1, FPF1, FLOWERING PROMOTING FACTOR 1 |
0.319 |
0.116 |
| 81 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.317 |
-1.492 |
| 82 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.317 |
-0.789 |
| 83 |
247800_at |
AT5G58570
|
unknown |
0.317 |
0.147 |
| 84 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.317 |
-0.040 |
| 85 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.317 |
-0.626 |
| 86 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.315 |
-0.407 |
| 87 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.314 |
-0.805 |
| 88 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.310 |
-0.352 |
| 89 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.308 |
0.277 |
| 90 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.306 |
0.481 |
| 91 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.306 |
-0.490 |
| 92 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.304 |
-0.198 |
| 93 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.302 |
-0.131 |
| 94 |
258203_at |
AT3G13950
|
unknown |
0.300 |
-0.269 |
| 95 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
0.299 |
-0.948 |
| 96 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.298 |
-0.525 |
| 97 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.293 |
-0.377 |
| 98 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.293 |
0.112 |
| 99 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.289 |
-0.815 |
| 100 |
263616_at |
AT2G04680
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.289 |
-1.445 |
| 101 |
260656_at |
AT1G19380
|
unknown |
0.287 |
-0.800 |
| 102 |
256631_at |
AT3G28320
|
unknown |
0.286 |
-0.748 |
| 103 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.286 |
-0.455 |
| 104 |
250659_at |
AT5G07050
|
UMAMIT9, Usually multiple acids move in and out Transporters 9 |
0.284 |
-0.859 |
| 105 |
247177_at |
AT5G65300
|
unknown |
0.284 |
-0.187 |
| 106 |
247751_at |
AT5G59050
|
unknown |
0.283 |
0.656 |
| 107 |
249013_at |
AT5G44700
|
EDA23, EMBRYO SAC DEVELOPMENT ARREST 23, GSO2, GASSHO 2 |
0.276 |
-0.251 |
| 108 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.276 |
-0.433 |
| 109 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
0.276 |
-1.095 |
| 110 |
260315_at |
AT1G63820
|
[CCT motif family protein] |
0.276 |
-0.798 |
| 111 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.273 |
0.280 |
| 112 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.271 |
-0.163 |
| 113 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.269 |
0.364 |
| 114 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.268 |
0.212 |
| 115 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.267 |
-0.761 |
| 116 |
264759_at |
AT1G61480
|
[S-locus lectin protein kinase family protein] |
0.267 |
-0.618 |
| 117 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
0.267 |
0.361 |
| 118 |
254632_at |
AT4G18630
|
unknown |
0.267 |
-1.312 |
| 119 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.266 |
-0.210 |
| 120 |
266486_at |
AT2G47950
|
unknown |
0.266 |
-1.018 |
| 121 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.265 |
-0.833 |
| 122 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.264 |
-0.964 |
| 123 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.264 |
-0.881 |
| 124 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.264 |
-0.936 |
| 125 |
246584_at |
AT5G14730
|
unknown |
0.263 |
-0.334 |
| 126 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.261 |
-0.327 |
| 127 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.261 |
-0.593 |
| 128 |
260858_at |
AT1G43770
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.259 |
0.827 |
| 129 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.259 |
-0.804 |
| 130 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
0.258 |
-0.518 |
| 131 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.258 |
-0.133 |
| 132 |
265276_at |
AT2G28400
|
unknown |
0.258 |
0.107 |
| 133 |
245713_at |
AT5G04370
|
NAMT1 |
0.258 |
-0.992 |
| 134 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.257 |
-0.166 |
| 135 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.256 |
-0.570 |
| 136 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.255 |
-0.492 |
| 137 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.255 |
-0.741 |
| 138 |
266396_at |
AT2G38790
|
unknown |
0.254 |
0.251 |
| 139 |
248343_at |
AT5G52260
|
AtMYB19, myb domain protein 19, MYB19, myb domain protein 19 |
0.254 |
-0.918 |
| 140 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.252 |
-0.378 |
| 141 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
0.252 |
-0.456 |
| 142 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
0.251 |
-0.434 |
| 143 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.250 |
0.695 |
| 144 |
266490_at |
AT2G07000
|
unknown |
0.249 |
-0.481 |
| 145 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.249 |
0.137 |
| 146 |
257815_at |
AT3G25130
|
unknown |
0.248 |
0.321 |
| 147 |
263081_at |
AT2G27220
|
BLH5, BEL1-like homeodomain 5 |
0.248 |
-0.283 |
| 148 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.247 |
-0.167 |
| 149 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
0.247 |
-0.639 |
| 150 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.246 |
-0.372 |
| 151 |
249797_at |
AT5G23750
|
[Remorin family protein] |
0.246 |
0.619 |
| 152 |
249534_at |
AT5G38800
|
AtbZIP43, basic leucine-zipper 43, bZIP43, basic leucine-zipper 43 |
0.245 |
-0.071 |
| 153 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.245 |
0.402 |
| 154 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.243 |
-0.095 |
| 155 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.241 |
-0.138 |
| 156 |
264151_at |
AT1G02070
|
unknown |
0.241 |
-0.172 |
| 157 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.241 |
-0.984 |
| 158 |
262873_at |
AT1G64700
|
unknown |
0.241 |
-0.555 |
| 159 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.240 |
-1.111 |
| 160 |
249622_at |
AT5G37550
|
unknown |
0.239 |
0.145 |
| 161 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.238 |
-0.131 |
| 162 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.237 |
-1.179 |
| 163 |
260635_at |
AT1G62422
|
unknown |
0.235 |
-0.145 |
| 164 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.234 |
-1.218 |
| 165 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.234 |
-0.506 |
| 166 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.234 |
-0.835 |
| 167 |
262138_at |
AT1G52660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.234 |
-1.099 |
| 168 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.233 |
-0.064 |
| 169 |
264119_at |
AT1G79180
|
ATMYB63, MYB DOMAIN PROTEIN 63, MYB63, myb domain protein 63 |
0.233 |
0.141 |
| 170 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.233 |
-0.565 |
| 171 |
259182_at |
AT3G01750
|
[Ankyrin repeat family protein] |
0.232 |
-0.011 |
| 172 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
0.232 |
-1.083 |
| 173 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.232 |
-0.452 |
| 174 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.231 |
-0.288 |
| 175 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.229 |
-0.135 |
| 176 |
250068_at |
AT5G17960
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.228 |
-1.029 |
| 177 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.227 |
0.610 |
| 178 |
259653_at |
AT1G55240
|
unknown |
0.227 |
-1.150 |
| 179 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.227 |
-0.323 |
| 180 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.226 |
-0.860 |
| 181 |
245194_at |
AT1G67820
|
[Protein phosphatase 2C family protein] |
0.225 |
0.162 |
| 182 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.225 |
-0.359 |
| 183 |
251058_at |
AT5G01790
|
unknown |
0.224 |
-0.404 |
| 184 |
265194_at |
AT1G05010
|
ACO4, ethylene forming enzyme, EAT1, EFE, ethylene-forming enzyme |
0.224 |
0.151 |
| 185 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.224 |
-0.701 |
| 186 |
256283_at |
AT3G12540
|
unknown |
0.224 |
-0.976 |
| 187 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
0.224 |
-0.517 |
| 188 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.223 |
0.137 |
| 189 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.223 |
-0.526 |
| 190 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
0.222 |
0.216 |
| 191 |
265460_at |
AT2G46600
|
[Calcium-binding EF-hand family protein] |
0.221 |
0.076 |
| 192 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
0.221 |
-0.223 |
| 193 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
0.220 |
-0.570 |
| 194 |
252505_at |
AT3G46170
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.220 |
-1.364 |
| 195 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.218 |
-0.992 |
| 196 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.218 |
-0.630 |
| 197 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.217 |
0.472 |
| 198 |
259991_at |
AT1G68040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.217 |
-0.592 |
| 199 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
0.217 |
-0.736 |
| 200 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.216 |
-0.690 |
| 201 |
259479_at |
AT1G19020
|
unknown |
0.216 |
-0.031 |
| 202 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.216 |
-0.103 |
| 203 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.215 |
-0.900 |
| 204 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
0.215 |
-1.298 |
| 205 |
247708_at |
AT5G59550
|
unknown |
0.215 |
0.783 |
| 206 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.214 |
-0.453 |
| 207 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.213 |
-0.848 |
| 208 |
258807_at |
AT3G04030
|
MYR2 |
0.213 |
-0.878 |
| 209 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.213 |
-0.640 |
| 210 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.212 |
-0.892 |
| 211 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.212 |
-0.535 |
| 212 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.212 |
-0.031 |
| 213 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.211 |
-0.920 |
| 214 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
0.211 |
-0.033 |
| 215 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.211 |
-0.051 |
| 216 |
257925_at |
AT3G23170
|
unknown |
0.210 |
0.262 |
| 217 |
264909_at |
AT2G17300
|
unknown |
0.210 |
-0.740 |
| 218 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.210 |
-0.051 |
| 219 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.208 |
0.461 |
| 220 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-1.068 |
1.115 |
| 221 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.895 |
1.229 |
| 222 |
250472_at |
AT5G10210
|
unknown |
-0.881 |
-0.944 |
| 223 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.851 |
-0.175 |
| 224 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.815 |
-0.316 |
| 225 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.757 |
-0.407 |
| 226 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.748 |
-0.424 |
| 227 |
252882_at |
AT4G39675
|
unknown |
-0.745 |
0.084 |
| 228 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.728 |
-0.059 |
| 229 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.707 |
0.053 |
| 230 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
-0.665 |
-0.029 |
| 231 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.663 |
-0.319 |
| 232 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.650 |
-1.193 |
| 233 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.645 |
-0.465 |
| 234 |
262399_at |
AT1G49500
|
unknown |
-0.613 |
0.569 |
| 235 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-0.611 |
-1.021 |
| 236 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.610 |
-0.829 |
| 237 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.583 |
0.557 |
| 238 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.581 |
-0.448 |
| 239 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.552 |
0.015 |
| 240 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.521 |
0.311 |
| 241 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.507 |
0.686 |
| 242 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.502 |
-0.436 |
| 243 |
266222_at |
AT2G28780
|
unknown |
-0.483 |
-0.431 |
| 244 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.480 |
-0.490 |
| 245 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.475 |
-0.454 |
| 246 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.475 |
-0.599 |
| 247 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
-0.472 |
-0.392 |
| 248 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.471 |
-0.656 |
| 249 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.468 |
-0.625 |
| 250 |
247754_at |
AT5G59080
|
unknown |
-0.468 |
-0.322 |
| 251 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.454 |
-0.850 |
| 252 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
-0.448 |
1.181 |
| 253 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.445 |
-0.433 |
| 254 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.435 |
-0.530 |
| 255 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.435 |
0.487 |
| 256 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
-0.432 |
-0.437 |
| 257 |
252501_at |
AT3G46880
|
unknown |
-0.426 |
-0.372 |
| 258 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.420 |
-0.471 |
| 259 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.418 |
-0.973 |
| 260 |
263285_at |
AT2G36120
|
DOT1, DEFECTIVELY ORGANIZED TRIBUTARIES 1 |
-0.417 |
-0.753 |
| 261 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.411 |
-0.780 |
| 262 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.401 |
-0.641 |
| 263 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.400 |
-0.639 |
| 264 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.395 |
0.486 |
| 265 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.392 |
-1.289 |
| 266 |
265481_at |
AT2G15960
|
unknown |
-0.391 |
0.057 |
| 267 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.391 |
-0.495 |
| 268 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.382 |
-0.700 |
| 269 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.378 |
-0.013 |
| 270 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.376 |
-0.111 |
| 271 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.376 |
0.395 |
| 272 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.374 |
0.828 |
| 273 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-0.370 |
0.986 |
| 274 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.368 |
0.382 |
| 275 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.366 |
0.242 |
| 276 |
265560_at |
AT2G05520
|
ATGRP-3, GLYCINE-RICH PROTEIN 3, ATGRP3, ARABIDOPSIS GLYCINE RICH PROTEIN 3, GRP-3, glycine-rich protein 3, GRP3, GLYCINE-RICH PROTEIN 3 |
-0.366 |
1.952 |
| 277 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
-0.362 |
-0.108 |
| 278 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.361 |
-0.804 |
| 279 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.358 |
-1.033 |
| 280 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.357 |
-0.760 |
| 281 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.355 |
0.473 |
| 282 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.353 |
-0.656 |
| 283 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.353 |
-0.802 |
| 284 |
256499_at |
AT1G36640
|
unknown |
-0.351 |
-0.935 |
| 285 |
261567_at |
AT1G33055
|
unknown |
-0.344 |
-0.016 |
| 286 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.340 |
1.247 |
| 287 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.333 |
0.859 |
| 288 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.332 |
-0.653 |
| 289 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.331 |
0.034 |
| 290 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.329 |
-1.211 |
| 291 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.328 |
-0.951 |
| 292 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.322 |
0.937 |
| 293 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.321 |
0.723 |
| 294 |
250828_at |
AT5G05250
|
unknown |
-0.320 |
-0.805 |
| 295 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
-0.320 |
0.560 |
| 296 |
246142_at |
AT5G19970
|
unknown |
-0.320 |
-0.031 |
| 297 |
247711_at |
AT5G59270
|
LecRK-II.2, L-type lectin receptor kinase II.2 |
-0.318 |
-1.351 |
| 298 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.317 |
-0.705 |
| 299 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.317 |
-0.153 |
| 300 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
-0.316 |
0.100 |
| 301 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.312 |
-0.512 |
| 302 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.311 |
0.351 |
| 303 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.311 |
0.029 |
| 304 |
257159_at |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
-0.309 |
-0.344 |
| 305 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
-0.304 |
-0.076 |
| 306 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
-0.303 |
-0.873 |
| 307 |
259996_at |
AT1G67910
|
unknown |
-0.302 |
-0.389 |
| 308 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.302 |
0.129 |
| 309 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.301 |
-0.165 |
| 310 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.301 |
0.555 |
| 311 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.299 |
0.377 |
| 312 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.298 |
0.011 |
| 313 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.298 |
0.524 |
| 314 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.298 |
-0.740 |
| 315 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.291 |
-0.920 |
| 316 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.291 |
0.046 |
| 317 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.291 |
0.617 |
| 318 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.290 |
1.057 |
| 319 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.290 |
-0.668 |
| 320 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.289 |
-0.577 |
| 321 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.289 |
0.856 |
| 322 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
-0.288 |
0.613 |
| 323 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.288 |
-0.142 |
| 324 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.287 |
-0.150 |
| 325 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.285 |
0.328 |
| 326 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.284 |
-0.741 |
| 327 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.283 |
-0.307 |
| 328 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
-0.282 |
-0.285 |
| 329 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.281 |
0.340 |
| 330 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.281 |
-0.917 |
| 331 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
-0.279 |
0.362 |
| 332 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.276 |
-0.674 |
| 333 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.275 |
0.331 |
| 334 |
255377_at |
AT4G03500
|
[Ankyrin repeat family protein] |
-0.274 |
-0.035 |
| 335 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.273 |
0.639 |
| 336 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
-0.273 |
0.394 |
| 337 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.273 |
-0.020 |
| 338 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.272 |
-0.487 |
| 339 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.271 |
-0.671 |
| 340 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
-0.270 |
0.470 |
| 341 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.269 |
0.392 |
| 342 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.267 |
-0.100 |
| 343 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.267 |
0.725 |
| 344 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.267 |
-0.813 |
| 345 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.266 |
-0.475 |
| 346 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.266 |
0.226 |
| 347 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.264 |
-0.920 |
| 348 |
248245_at |
AT5G53190
|
AtSWEET3, SWEET3 |
-0.261 |
-0.800 |
| 349 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
-0.259 |
1.473 |
| 350 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.258 |
0.704 |
| 351 |
260495_at |
AT2G41810
|
unknown |
-0.257 |
-0.847 |
| 352 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.256 |
0.598 |
| 353 |
249136_at |
AT5G43180
|
unknown |
-0.255 |
0.231 |
| 354 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.254 |
-1.203 |
| 355 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.253 |
-0.469 |
| 356 |
249932_at |
AT5G22390
|
unknown |
-0.252 |
-0.017 |
| 357 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-0.250 |
0.630 |
| 358 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
-0.248 |
0.521 |
| 359 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.248 |
0.770 |
| 360 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.248 |
0.107 |
| 361 |
265186_at |
AT1G23560
|
unknown |
-0.247 |
-0.882 |
| 362 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.246 |
0.587 |
| 363 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.244 |
0.184 |
| 364 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.244 |
0.003 |
| 365 |
260668_at |
AT1G19530
|
unknown |
-0.243 |
-0.327 |
| 366 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.243 |
1.169 |
| 367 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.242 |
0.360 |
| 368 |
249635_at |
AT5G36870
|
ATGSL09, glucan synthase-like 9, atgsl9, gsl09, GSL09, glucan synthase-like 9 |
-0.242 |
0.325 |
| 369 |
265897_at |
AT2G25680
|
MOT1, molybdate transporter 1 |
-0.240 |
-0.438 |
| 370 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
-0.240 |
1.469 |
| 371 |
253317_at |
AT4G33960
|
unknown |
-0.240 |
-0.341 |
| 372 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.239 |
0.773 |
| 373 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
-0.238 |
1.757 |
| 374 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.237 |
-0.471 |
| 375 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.237 |
1.102 |
| 376 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.235 |
0.438 |
| 377 |
245076_at |
AT2G23170
|
GH3.3 |
-0.235 |
0.572 |
| 378 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
-0.234 |
-0.496 |
| 379 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.234 |
1.083 |
| 380 |
263481_at |
AT2G04025
|
GLV7, GOLVEN 7, RGF3, root meristem growth factor 3 |
-0.233 |
-0.593 |
| 381 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
-0.232 |
-0.437 |
| 382 |
256847_at |
AT3G27950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.232 |
-0.703 |
| 383 |
259316_at |
AT3G01175
|
unknown |
-0.232 |
-1.197 |
| 384 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.231 |
-0.633 |
| 385 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.230 |
-0.930 |
| 386 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.230 |
0.188 |
| 387 |
263805_at |
AT2G40400
|
unknown |
-0.229 |
-0.431 |
| 388 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
-0.228 |
0.052 |
| 389 |
245026_at |
ATCG00140
|
ATPH |
-0.226 |
0.533 |
| 390 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.226 |
-0.586 |
| 391 |
250217_at |
AT5G14120
|
[Major facilitator superfamily protein] |
-0.225 |
0.278 |
| 392 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-0.225 |
-0.731 |
| 393 |
248827_at |
AT5G47100
|
ATCBL9, CBL9, calcineurin B-like protein 9 |
-0.225 |
0.431 |
| 394 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.225 |
0.042 |
| 395 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.222 |
1.309 |
| 396 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.222 |
-0.055 |
| 397 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.220 |
-0.768 |
| 398 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
-0.220 |
0.508 |
| 399 |
258370_at |
AT3G14395
|
unknown |
-0.220 |
-0.447 |
| 400 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.219 |
-0.571 |
| 401 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
-0.219 |
-0.144 |
| 402 |
256799_at |
AT3G18560
|
unknown |
-0.218 |
0.192 |
| 403 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.216 |
-0.332 |
| 404 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.216 |
-1.059 |
| 405 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
-0.216 |
-0.571 |
| 406 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
-0.215 |
-0.040 |
| 407 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
-0.213 |
0.173 |
| 408 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.211 |
0.642 |
| 409 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.211 |
0.312 |
| 410 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.211 |
0.266 |
| 411 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.211 |
0.939 |
| 412 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.210 |
1.479 |
| 413 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.208 |
0.138 |
| 414 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
-0.208 |
-0.079 |
| 415 |
247649_at |
AT5G60030
|
unknown |
-0.207 |
-0.233 |
| 416 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.206 |
0.952 |
| 417 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
-0.204 |
0.396 |
| 418 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.203 |
-0.309 |
| 419 |
260771_at |
AT1G49160
|
WNK7 |
-0.202 |
-1.290 |
| 420 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.201 |
-0.539 |
| 421 |
252116_at |
AT3G51510
|
unknown |
-0.201 |
0.071 |
| 422 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.201 |
-0.618 |
| 423 |
256796_at |
AT3G22210
|
unknown |
-0.201 |
0.794 |
| 424 |
250223_at |
AT5G14070
|
ROXY2 |
-0.200 |
-0.345 |
| 425 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.200 |
-1.140 |
| 426 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.199 |
-0.035 |
| 427 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
-0.199 |
-1.148 |
| 428 |
249766_at |
AT5G24070
|
[Peroxidase superfamily protein] |
-0.199 |
0.190 |
| 429 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.199 |
1.316 |
| 430 |
262172_at |
AT1G74970
|
RPS9, ribosomal protein S9, TWN3 |
-0.199 |
-0.458 |
| 431 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
-0.199 |
-1.206 |
| 432 |
262396_at |
AT1G49470
|
unknown |
-0.197 |
0.560 |
| 433 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.197 |
-1.238 |
| 434 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.195 |
-0.230 |
| 435 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.195 |
0.026 |
| 436 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.195 |
-0.488 |
| 437 |
245547_at |
AT4G15300
|
CYP702A2, cytochrome P450, family 702, subfamily A, polypeptide 2 |
-0.194 |
-1.215 |
| 438 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.192 |
-0.056 |