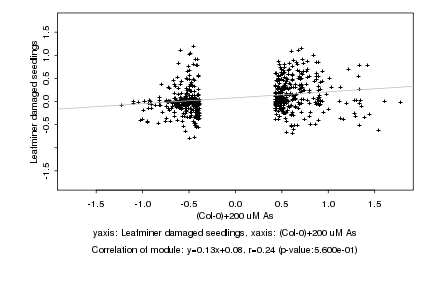
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
(Col-0)+200 uM As |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.774 |
-0.005 |
| 2 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.603 |
0.006 |
| 3 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.533 |
-0.611 |
| 4 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.434 |
-0.272 |
| 5 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
1.421 |
0.786 |
| 6 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
1.386 |
-0.340 |
| 7 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.352 |
-0.105 |
| 8 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
1.344 |
0.034 |
| 9 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
1.335 |
0.280 |
| 10 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.331 |
0.797 |
| 11 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.331 |
-0.517 |
| 12 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.326 |
-0.030 |
| 13 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
1.323 |
0.553 |
| 14 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
1.297 |
0.027 |
| 15 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.286 |
-0.321 |
| 16 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.281 |
0.035 |
| 17 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.276 |
-0.256 |
| 18 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
1.218 |
0.701 |
| 19 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.190 |
-0.022 |
| 20 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.159 |
-0.371 |
| 21 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
1.128 |
0.314 |
| 22 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
1.122 |
-0.358 |
| 23 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.115 |
0.015 |
| 24 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
1.028 |
0.308 |
| 25 |
253044_at |
AT4G37290
|
unknown |
1.004 |
0.512 |
| 26 |
254263_at |
AT4G23493
|
unknown |
0.994 |
-0.172 |
| 27 |
245082_at |
AT2G23270
|
unknown |
0.977 |
0.172 |
| 28 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.947 |
-0.217 |
| 29 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.937 |
0.001 |
| 30 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.937 |
0.653 |
| 31 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.931 |
0.443 |
| 32 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.916 |
0.421 |
| 33 |
250624_at |
AT5G07330
|
unknown |
0.914 |
0.011 |
| 34 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.913 |
0.280 |
| 35 |
258827_at |
AT3G07150
|
unknown |
0.911 |
-0.051 |
| 36 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.907 |
0.024 |
| 37 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.902 |
0.090 |
| 38 |
263515_at |
AT2G21640
|
unknown |
0.898 |
0.136 |
| 39 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.895 |
-0.070 |
| 40 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.892 |
0.864 |
| 41 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.892 |
-0.396 |
| 42 |
245272_at |
AT4G17250
|
unknown |
0.891 |
0.015 |
| 43 |
245243_at |
AT1G44414
|
unknown |
0.889 |
0.108 |
| 44 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.889 |
0.329 |
| 45 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.886 |
0.559 |
| 46 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.886 |
-0.039 |
| 47 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.885 |
0.251 |
| 48 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
0.883 |
0.052 |
| 49 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.869 |
0.559 |
| 50 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.868 |
-0.063 |
| 51 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.866 |
0.611 |
| 52 |
254536_at |
AT4G19720
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
0.864 |
-0.045 |
| 53 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.864 |
0.477 |
| 54 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.863 |
0.852 |
| 55 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.862 |
-0.489 |
| 56 |
248316_at |
AT5G52670
|
[Copper transport protein family] |
0.860 |
-0.013 |
| 57 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.849 |
0.445 |
| 58 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.833 |
1.003 |
| 59 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.833 |
0.082 |
| 60 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.804 |
-0.480 |
| 61 |
252946_at |
AT4G39235
|
unknown |
0.799 |
-0.235 |
| 62 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.795 |
-0.313 |
| 63 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.794 |
-0.035 |
| 64 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.791 |
0.208 |
| 65 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.791 |
0.559 |
| 66 |
250826_at |
AT5G05220
|
unknown |
0.788 |
0.239 |
| 67 |
255950_at |
AT1G22110
|
[structural constituent of ribosome] |
0.776 |
0.030 |
| 68 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.775 |
0.533 |
| 69 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.775 |
0.693 |
| 70 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.771 |
0.866 |
| 71 |
251950_at |
AT3G53600
|
[C2H2-type zinc finger family protein] |
0.761 |
0.663 |
| 72 |
255485_at |
AT4G02550
|
unknown |
0.757 |
0.378 |
| 73 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.741 |
0.248 |
| 74 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.735 |
-0.373 |
| 75 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.730 |
0.060 |
| 76 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.728 |
0.833 |
| 77 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.727 |
0.260 |
| 78 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.726 |
0.654 |
| 79 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.715 |
0.307 |
| 80 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.713 |
0.597 |
| 81 |
253827_at |
AT4G28085
|
unknown |
0.712 |
0.924 |
| 82 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.706 |
0.362 |
| 83 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.706 |
-0.037 |
| 84 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.705 |
0.234 |
| 85 |
260047_at |
AT1G73740
|
[UDP-Glycosyltransferase superfamily protein] |
0.704 |
0.064 |
| 86 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.704 |
-0.227 |
| 87 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.702 |
1.153 |
| 88 |
246018_at |
AT5G10695
|
unknown |
0.702 |
0.515 |
| 89 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.702 |
0.371 |
| 90 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.702 |
0.625 |
| 91 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.699 |
0.203 |
| 92 |
258939_at |
AT3G10020
|
unknown |
0.698 |
0.296 |
| 93 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.696 |
0.498 |
| 94 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.692 |
0.499 |
| 95 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.690 |
0.543 |
| 96 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.690 |
0.355 |
| 97 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.689 |
0.515 |
| 98 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
0.686 |
-0.047 |
| 99 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.684 |
0.658 |
| 100 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.680 |
0.262 |
| 101 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.677 |
0.509 |
| 102 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.674 |
-0.139 |
| 103 |
249618_at |
AT5G37490
|
[ARM repeat superfamily protein] |
0.673 |
-0.010 |
| 104 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.669 |
1.122 |
| 105 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.668 |
0.677 |
| 106 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.666 |
-0.505 |
| 107 |
245662_at |
AT1G28190
|
unknown |
0.660 |
0.806 |
| 108 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.653 |
0.162 |
| 109 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.652 |
0.856 |
| 110 |
260584_at |
AT2G43660
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.646 |
0.050 |
| 111 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.646 |
-0.110 |
| 112 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.645 |
0.322 |
| 113 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.645 |
0.142 |
| 114 |
249896_at |
AT5G22530
|
unknown |
0.643 |
0.375 |
| 115 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.642 |
-0.105 |
| 116 |
258364_at |
AT3G14225
|
GLIP4, GDSL-motif lipase 4 |
0.641 |
0.084 |
| 117 |
253796_at |
AT4G28460
|
unknown |
0.632 |
0.082 |
| 118 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.628 |
-0.516 |
| 119 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.623 |
0.902 |
| 120 |
245755_at |
AT1G35210
|
unknown |
0.622 |
0.313 |
| 121 |
247287_at |
AT5G64230
|
unknown |
0.621 |
0.057 |
| 122 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
0.621 |
0.126 |
| 123 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.616 |
-0.588 |
| 124 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
0.615 |
-0.288 |
| 125 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.614 |
0.566 |
| 126 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.613 |
0.496 |
| 127 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.613 |
-0.687 |
| 128 |
253302_at |
AT4G33660
|
unknown |
0.609 |
-0.221 |
| 129 |
260495_at |
AT2G41810
|
unknown |
0.609 |
0.055 |
| 130 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.608 |
0.461 |
| 131 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.607 |
0.162 |
| 132 |
260005_at |
AT1G67920
|
unknown |
0.607 |
0.768 |
| 133 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.605 |
0.592 |
| 134 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.604 |
0.025 |
| 135 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.603 |
-0.503 |
| 136 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.603 |
1.085 |
| 137 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.601 |
0.312 |
| 138 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.600 |
-0.272 |
| 139 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.599 |
-0.525 |
| 140 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
0.598 |
-0.282 |
| 141 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.595 |
-0.261 |
| 142 |
248774_at |
AT5G47830
|
unknown |
0.594 |
-0.070 |
| 143 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.590 |
0.186 |
| 144 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.589 |
0.387 |
| 145 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.583 |
-0.092 |
| 146 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.580 |
0.136 |
| 147 |
260411_at |
AT1G69890
|
unknown |
0.576 |
0.696 |
| 148 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.575 |
0.840 |
| 149 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.574 |
0.292 |
| 150 |
260872_at |
AT1G21350
|
[Thioredoxin superfamily protein] |
0.573 |
-0.112 |
| 151 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.573 |
0.407 |
| 152 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.571 |
0.279 |
| 153 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.571 |
0.596 |
| 154 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.567 |
0.672 |
| 155 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
0.566 |
-0.097 |
| 156 |
246612_at |
AT5G35320
|
unknown |
0.565 |
0.002 |
| 157 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.564 |
0.594 |
| 158 |
263566_at |
AT2G15340
|
[glycine-rich protein] |
0.560 |
0.028 |
| 159 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.559 |
0.496 |
| 160 |
254715_at |
AT4G13550
|
[triglyceride lipases] |
0.556 |
-0.180 |
| 161 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.553 |
0.291 |
| 162 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.550 |
-0.424 |
| 163 |
264726_at |
AT1G22985
|
CRF7, cytokinin response factor 7 |
0.548 |
0.091 |
| 164 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.547 |
0.151 |
| 165 |
260833_at |
AT1G06800
|
DALL4, DAD1-Like Lipase 4, PLA-I{gamma}1, phospholipase A I gamma 1 |
0.546 |
0.138 |
| 166 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.544 |
0.501 |
| 167 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.544 |
-0.221 |
| 168 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.544 |
0.181 |
| 169 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.543 |
0.445 |
| 170 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.543 |
0.162 |
| 171 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.541 |
-0.650 |
| 172 |
249824_at |
AT5G23380
|
unknown |
0.540 |
-0.031 |
| 173 |
246370_at |
AT1G51920
|
unknown |
0.538 |
0.364 |
| 174 |
256518_at |
AT1G66080
|
unknown |
0.536 |
-0.351 |
| 175 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.535 |
-0.020 |
| 176 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.535 |
0.392 |
| 177 |
257026_at |
AT3G19200
|
unknown |
0.532 |
0.013 |
| 178 |
262014_at |
AT1G35660
|
unknown |
0.530 |
-0.257 |
| 179 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.529 |
0.320 |
| 180 |
249256_at |
AT5G41620
|
unknown |
0.529 |
0.057 |
| 181 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.527 |
0.043 |
| 182 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.525 |
0.806 |
| 183 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.525 |
0.262 |
| 184 |
263475_at |
AT2G31945
|
unknown |
0.523 |
0.527 |
| 185 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.522 |
0.327 |
| 186 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.520 |
-0.241 |
| 187 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.520 |
0.361 |
| 188 |
248717_at |
AT5G48175
|
unknown |
0.519 |
-0.037 |
| 189 |
250013_at |
AT5G18040
|
unknown |
0.518 |
-0.035 |
| 190 |
261748_at |
AT1G76070
|
unknown |
0.518 |
0.644 |
| 191 |
265618_at |
AT2G25460
|
unknown |
0.518 |
0.749 |
| 192 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.516 |
-0.228 |
| 193 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.516 |
0.023 |
| 194 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.516 |
0.272 |
| 195 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
0.515 |
-0.113 |
| 196 |
256499_at |
AT1G36640
|
unknown |
0.515 |
0.084 |
| 197 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.513 |
-0.241 |
| 198 |
264972_at |
AT1G67370
|
ASY1, ASYNAPTIC 1, ATASY1 |
0.513 |
-0.015 |
| 199 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
0.511 |
-0.214 |
| 200 |
252400_at |
AT3G48020
|
unknown |
0.511 |
0.463 |
| 201 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.510 |
0.097 |
| 202 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.509 |
-0.080 |
| 203 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.509 |
0.676 |
| 204 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.508 |
0.615 |
| 205 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.508 |
0.333 |
| 206 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.507 |
-0.149 |
| 207 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.505 |
0.005 |
| 208 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
0.505 |
0.115 |
| 209 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.504 |
0.001 |
| 210 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.504 |
0.595 |
| 211 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.503 |
0.256 |
| 212 |
253637_at |
AT4G30390
|
unknown |
0.502 |
0.209 |
| 213 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.501 |
0.318 |
| 214 |
255681_at |
AT4G00550
|
DGD2, digalactosyl diacylglycerol deficient 2 |
0.501 |
0.047 |
| 215 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.498 |
-0.154 |
| 216 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.496 |
0.497 |
| 217 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.495 |
0.207 |
| 218 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.495 |
0.285 |
| 219 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.493 |
0.643 |
| 220 |
257874_at |
AT3G17110
|
[pseudogene] |
0.492 |
0.268 |
| 221 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.492 |
-0.190 |
| 222 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.492 |
0.701 |
| 223 |
251558_at |
AT3G57810
|
[Cysteine proteinases superfamily protein] |
0.491 |
-0.161 |
| 224 |
257118_at |
AT3G20180
|
[Copper transport protein family] |
0.488 |
-0.069 |
| 225 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.488 |
0.105 |
| 226 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
0.488 |
-0.013 |
| 227 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.486 |
0.489 |
| 228 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.486 |
0.634 |
| 229 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.486 |
0.073 |
| 230 |
259923_at |
AT1G72760
|
[Protein kinase superfamily protein] |
0.485 |
-0.027 |
| 231 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
0.484 |
-0.136 |
| 232 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.483 |
0.001 |
| 233 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.482 |
0.030 |
| 234 |
263541_at |
AT2G24860
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
0.482 |
0.030 |
| 235 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
0.480 |
0.223 |
| 236 |
245200_at |
AT1G67850
|
unknown |
0.480 |
0.140 |
| 237 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.478 |
0.748 |
| 238 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.478 |
0.183 |
| 239 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.478 |
0.573 |
| 240 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.473 |
-0.312 |
| 241 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.473 |
0.811 |
| 242 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
0.472 |
-0.009 |
| 243 |
265077_at |
AT1G55530
|
[RING/U-box superfamily protein] |
0.472 |
-0.141 |
| 244 |
257925_at |
AT3G23170
|
unknown |
0.471 |
0.469 |
| 245 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.471 |
0.120 |
| 246 |
263897_at |
AT2G21940
|
ATSK1, SK1, shikimate kinase 1 |
0.470 |
-0.098 |
| 247 |
264465_at |
AT1G10230
|
ASK18, SKP1-like 18, SK18, SKP1-like 18 |
0.469 |
0.005 |
| 248 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.468 |
0.063 |
| 249 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.467 |
-0.323 |
| 250 |
263881_at |
AT2G21820
|
unknown |
0.466 |
0.408 |
| 251 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.466 |
0.288 |
| 252 |
249747_at |
AT5G24600
|
unknown |
0.464 |
0.085 |
| 253 |
261839_at |
AT1G16040
|
unknown |
0.463 |
0.117 |
| 254 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.461 |
0.328 |
| 255 |
247899_at |
AT5G57345
|
unknown |
0.461 |
-0.002 |
| 256 |
245254_at |
AT4G14680
|
APS3 |
0.461 |
-0.010 |
| 257 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.458 |
0.557 |
| 258 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.458 |
0.213 |
| 259 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.458 |
-0.081 |
| 260 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.457 |
0.568 |
| 261 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
0.456 |
-0.382 |
| 262 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
0.456 |
-0.027 |
| 263 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
0.455 |
-0.201 |
| 264 |
254408_at |
AT4G21390
|
B120 |
0.455 |
0.882 |
| 265 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
0.454 |
0.097 |
| 266 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.454 |
0.517 |
| 267 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.453 |
0.050 |
| 268 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.449 |
0.270 |
| 269 |
259979_at |
AT1G76600
|
unknown |
0.449 |
0.435 |
| 270 |
259027_at |
AT3G09280
|
unknown |
0.449 |
-0.055 |
| 271 |
246989_at |
AT5G67350
|
unknown |
0.449 |
-0.001 |
| 272 |
255077_at |
AT4G09150
|
[T-complex protein 11] |
0.447 |
-0.078 |
| 273 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.447 |
0.468 |
| 274 |
245051_at |
AT2G23320
|
AtWRKY15, WRKY15, WRKY DNA-binding protein 15 |
0.447 |
0.302 |
| 275 |
258262_at |
AT3G15770
|
unknown |
0.442 |
0.063 |
| 276 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.442 |
-0.089 |
| 277 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.440 |
0.621 |
| 278 |
256331_at |
AT1G76880
|
[Duplicated homeodomain-like superfamily protein] |
0.439 |
-0.274 |
| 279 |
252539_at |
AT3G45730
|
unknown |
0.437 |
0.112 |
| 280 |
254211_at |
AT4G23570
|
SGT1A |
0.436 |
-0.131 |
| 281 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.436 |
0.186 |
| 282 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.435 |
0.475 |
| 283 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.431 |
0.136 |
| 284 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.430 |
-0.081 |
| 285 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.429 |
-0.004 |
| 286 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.428 |
0.176 |
| 287 |
253913_at |
AT4G27370
|
ATVIIIB, MYOSIN VIII B, VIIIB |
0.427 |
-0.143 |
| 288 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.427 |
0.622 |
| 289 |
257421_at |
AT1G12030
|
unknown |
0.427 |
-0.375 |
| 290 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.427 |
0.351 |
| 291 |
246158_at |
AT5G19855
|
AtRbcX2, RbcX2, homologue of cyanobacterial RbcX 2 |
0.426 |
-0.029 |
| 292 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
0.426 |
0.063 |
| 293 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
0.426 |
-0.184 |
| 294 |
250925_at |
AT5G03370
|
[acylphosphatase family] |
0.426 |
-0.055 |
| 295 |
246125_at |
AT5G19875
|
unknown |
0.425 |
0.008 |
| 296 |
267537_at |
AT2G41880
|
AGK1, GK-1, guanylate kinase 1 |
0.424 |
0.044 |
| 297 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.424 |
-0.136 |
| 298 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.424 |
0.339 |
| 299 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
0.424 |
0.212 |
| 300 |
259019_at |
AT3G07370
|
ATCHIP, CARBOXYL TERMINUS OF HSC70-INTERACTING PROTEIN, CHIP, carboxyl terminus of HSC70-interacting protein |
0.423 |
0.111 |
| 301 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-1.234 |
-0.076 |
| 302 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-1.105 |
0.021 |
| 303 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-1.101 |
-0.036 |
| 304 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-1.052 |
-0.019 |
| 305 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-1.023 |
-0.401 |
| 306 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.011 |
-0.383 |
| 307 |
246142_at |
AT5G19970
|
unknown |
-0.994 |
-0.183 |
| 308 |
266222_at |
AT2G28780
|
unknown |
-0.988 |
0.005 |
| 309 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.982 |
0.016 |
| 310 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.956 |
-0.411 |
| 311 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.952 |
-0.448 |
| 312 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.946 |
-0.147 |
| 313 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.944 |
-0.035 |
| 314 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.926 |
0.029 |
| 315 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
-0.916 |
0.009 |
| 316 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.915 |
-0.141 |
| 317 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.913 |
-0.063 |
| 318 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.872 |
-0.079 |
| 319 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.837 |
-0.467 |
| 320 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.826 |
0.029 |
| 321 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.824 |
-0.000 |
| 322 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.822 |
0.092 |
| 323 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.821 |
-0.068 |
| 324 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.812 |
-0.068 |
| 325 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.799 |
0.381 |
| 326 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.796 |
-0.216 |
| 327 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.756 |
-0.425 |
| 328 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.750 |
0.033 |
| 329 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
-0.750 |
0.004 |
| 330 |
263545_at |
AT2G21560
|
unknown |
-0.746 |
-0.365 |
| 331 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.746 |
-0.091 |
| 332 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.744 |
0.013 |
| 333 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.733 |
-0.019 |
| 334 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.729 |
-0.137 |
| 335 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.727 |
-0.020 |
| 336 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.727 |
0.015 |
| 337 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.724 |
-0.004 |
| 338 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.723 |
0.021 |
| 339 |
247912_at |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.723 |
0.317 |
| 340 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.721 |
0.013 |
| 341 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.715 |
0.286 |
| 342 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.713 |
-0.185 |
| 343 |
250438_at |
AT5G10580
|
unknown |
-0.704 |
0.015 |
| 344 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.702 |
-0.416 |
| 345 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
-0.699 |
-0.137 |
| 346 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.697 |
-0.011 |
| 347 |
263596_at |
AT2G01900
|
[DNAse I-like superfamily protein] |
-0.687 |
-0.022 |
| 348 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.683 |
-0.091 |
| 349 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.675 |
-0.021 |
| 350 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.670 |
0.473 |
| 351 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.666 |
0.371 |
| 352 |
252568_at |
AT3G45410
|
LecRK-I.3, L-type lectin receptor kinase I.3 |
-0.663 |
0.075 |
| 353 |
264794_at |
AT1G08670
|
[ENTH/VHS family protein] |
-0.661 |
-0.023 |
| 354 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.661 |
-0.227 |
| 355 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.660 |
0.183 |
| 356 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.658 |
-0.305 |
| 357 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.657 |
-0.222 |
| 358 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.653 |
0.339 |
| 359 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.652 |
-0.080 |
| 360 |
252882_at |
AT4G39675
|
unknown |
-0.648 |
0.080 |
| 361 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
-0.646 |
0.013 |
| 362 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.645 |
-0.113 |
| 363 |
259596_at |
AT1G28130
|
GH3.17 |
-0.637 |
-0.044 |
| 364 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.635 |
0.037 |
| 365 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.635 |
-0.073 |
| 366 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.635 |
-0.003 |
| 367 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.634 |
0.520 |
| 368 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.633 |
-0.405 |
| 369 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
-0.630 |
0.020 |
| 370 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.629 |
-0.062 |
| 371 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
-0.629 |
-0.088 |
| 372 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.628 |
-0.036 |
| 373 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.624 |
0.826 |
| 374 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.619 |
0.156 |
| 375 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.614 |
-0.100 |
| 376 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
-0.614 |
0.301 |
| 377 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.612 |
-0.018 |
| 378 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.610 |
0.037 |
| 379 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.604 |
-0.131 |
| 380 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.603 |
-0.062 |
| 381 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.602 |
-0.123 |
| 382 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.599 |
0.012 |
| 383 |
253832_at |
AT4G27654
|
unknown |
-0.598 |
1.119 |
| 384 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.597 |
0.034 |
| 385 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.596 |
0.369 |
| 386 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.594 |
0.108 |
| 387 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.592 |
0.014 |
| 388 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.590 |
-0.163 |
| 389 |
245554_at |
AT4G15380
|
CYP705A4, cytochrome P450, family 705, subfamily A, polypeptide 4 |
-0.590 |
0.002 |
| 390 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.589 |
0.446 |
| 391 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.586 |
-0.083 |
| 392 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.586 |
0.081 |
| 393 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.586 |
-0.044 |
| 394 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.585 |
-0.050 |
| 395 |
246374_at |
AT1G51840
|
[protein kinase-related] |
-0.585 |
-0.021 |
| 396 |
262399_at |
AT1G49500
|
unknown |
-0.582 |
0.187 |
| 397 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.581 |
-0.142 |
| 398 |
262396_at |
AT1G49470
|
unknown |
-0.577 |
-0.011 |
| 399 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.577 |
-0.318 |
| 400 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
-0.575 |
-0.424 |
| 401 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.571 |
0.002 |
| 402 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.571 |
0.050 |
| 403 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.569 |
-0.008 |
| 404 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.567 |
-0.056 |
| 405 |
245007_at |
ATCG00350
|
PSAA |
-0.566 |
-0.070 |
| 406 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.564 |
0.025 |
| 407 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.563 |
-0.067 |
| 408 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.562 |
-0.077 |
| 409 |
258017_at |
AT3G19370
|
unknown |
-0.561 |
-0.532 |
| 410 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.560 |
-0.239 |
| 411 |
256684_at |
AT3G32040
|
[Terpenoid synthases superfamily protein] |
-0.557 |
0.083 |
| 412 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.557 |
-0.311 |
| 413 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.553 |
0.058 |
| 414 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.551 |
0.025 |
| 415 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.549 |
-0.034 |
| 416 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.547 |
0.115 |
| 417 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.547 |
-0.147 |
| 418 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.546 |
0.103 |
| 419 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.543 |
-0.252 |
| 420 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
-0.542 |
-0.087 |
| 421 |
246001_at |
AT5G20790
|
unknown |
-0.542 |
-0.646 |
| 422 |
262863_at |
AT1G64910
|
[UDP-Glycosyltransferase superfamily protein] |
-0.542 |
0.083 |
| 423 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.538 |
0.165 |
| 424 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.538 |
-0.007 |
| 425 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.537 |
0.025 |
| 426 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.536 |
0.113 |
| 427 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.533 |
-0.076 |
| 428 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.533 |
0.298 |
| 429 |
253989_at |
AT4G26130
|
unknown |
-0.530 |
0.190 |
| 430 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.530 |
0.514 |
| 431 |
244997_at |
ATCG00170
|
RPOC2 |
-0.528 |
-0.174 |
| 432 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.528 |
-0.210 |
| 433 |
244962_at |
ATCG01050
|
NDHD |
-0.528 |
-0.093 |
| 434 |
244935_at |
ATCG01090
|
NDHI |
-0.525 |
-0.049 |
| 435 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.525 |
0.005 |
| 436 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.524 |
0.241 |
| 437 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.523 |
0.016 |
| 438 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.523 |
0.044 |
| 439 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.521 |
0.494 |
| 440 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.521 |
0.653 |
| 441 |
247754_at |
AT5G59080
|
unknown |
-0.521 |
0.178 |
| 442 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.519 |
-0.343 |
| 443 |
260332_at |
AT1G70470
|
unknown |
-0.519 |
-0.073 |
| 444 |
262452_at |
AT1G11210
|
unknown |
-0.516 |
0.762 |
| 445 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.516 |
-0.037 |
| 446 |
256531_at |
AT1G33320
|
[Pyridoxal phosphate (PLP)-dependent transferases superfamily protein] |
-0.515 |
-0.006 |
| 447 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.515 |
-0.009 |
| 448 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.515 |
-0.040 |
| 449 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.514 |
0.003 |
| 450 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.511 |
0.014 |
| 451 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.508 |
-0.146 |
| 452 |
245025_at |
ATCG00130
|
ATPF |
-0.506 |
-0.147 |
| 453 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.505 |
0.134 |
| 454 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.505 |
0.265 |
| 455 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.503 |
0.687 |
| 456 |
245776_at |
AT1G30260
|
unknown |
-0.503 |
0.329 |
| 457 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.502 |
-0.783 |
| 458 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.501 |
-0.037 |
| 459 |
250450_at |
AT5G10280
|
ATMYB64, ATMYB92, myb domain protein 92, MYB92, myb domain protein 92 |
-0.500 |
-0.013 |
| 460 |
265057_at |
AT1G52140
|
unknown |
-0.499 |
0.069 |
| 461 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.498 |
-0.342 |
| 462 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.498 |
0.478 |
| 463 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
-0.497 |
-0.061 |
| 464 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.497 |
0.046 |
| 465 |
267217_at |
AT2G02610
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.496 |
-0.025 |
| 466 |
263032_at |
AT1G23850
|
unknown |
-0.496 |
0.580 |
| 467 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.495 |
1.018 |
| 468 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.493 |
0.386 |
| 469 |
259887_at |
AT1G76360
|
[Protein kinase superfamily protein] |
-0.492 |
0.224 |
| 470 |
261090_at |
AT1G07560
|
[Leucine-rich repeat protein kinase family protein] |
-0.492 |
-0.022 |
| 471 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.490 |
-0.012 |
| 472 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.490 |
-0.228 |
| 473 |
246580_at |
AT1G31770
|
ABCG14, ATP-binding cassette G14 |
-0.490 |
-0.148 |
| 474 |
253556_at |
AT4G31100
|
[wall-associated kinase, putative] |
-0.490 |
-0.030 |
| 475 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.488 |
1.055 |
| 476 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.487 |
-0.327 |
| 477 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.484 |
-0.281 |
| 478 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.483 |
0.221 |
| 479 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.481 |
-0.003 |
| 480 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
-0.480 |
-0.168 |
| 481 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.479 |
0.091 |
| 482 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.478 |
-0.224 |
| 483 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.475 |
0.432 |
| 484 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
-0.473 |
0.345 |
| 485 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.471 |
0.023 |
| 486 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.466 |
0.354 |
| 487 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.466 |
0.143 |
| 488 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.464 |
0.077 |
| 489 |
249687_at |
AT5G36150
|
ATPEN3, putative pentacyclic triterpene synthase 3, PEN3, putative pentacyclic triterpene synthase 3 |
-0.464 |
0.044 |
| 490 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.462 |
-0.060 |
| 491 |
257299_at |
AT3G28050
|
UMAMIT41, Usually multiple acids move in and out Transporters 41 |
-0.462 |
-0.094 |
| 492 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-0.462 |
1.204 |
| 493 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.462 |
0.802 |
| 494 |
261946_at |
AT1G64560
|
|
-0.461 |
0.025 |
| 495 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.460 |
0.005 |
| 496 |
250472_at |
AT5G10210
|
unknown |
-0.459 |
-0.029 |
| 497 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.459 |
0.478 |
| 498 |
259017_at |
AT3G07310
|
unknown |
-0.458 |
-0.179 |
| 499 |
244936_at |
ATCG01100
|
NDHA |
-0.458 |
-0.072 |
| 500 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.456 |
-0.038 |
| 501 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.456 |
-0.414 |
| 502 |
255608_at |
AT4G01140
|
unknown |
-0.454 |
-0.013 |
| 503 |
263146_at |
AT1G53940
|
GLIP2, GDSL-motif lipase 2 |
-0.453 |
-0.023 |
| 504 |
247070_at |
AT5G66815
|
unknown |
-0.451 |
0.005 |
| 505 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.450 |
0.069 |
| 506 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
-0.449 |
0.071 |
| 507 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.449 |
0.029 |
| 508 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.449 |
-0.455 |
| 509 |
261500_at |
AT1G28400
|
unknown |
-0.448 |
0.100 |
| 510 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.447 |
0.038 |
| 511 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.447 |
0.081 |
| 512 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.447 |
-0.048 |
| 513 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.445 |
0.223 |
| 514 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.445 |
0.325 |
| 515 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.445 |
0.469 |
| 516 |
245006_at |
ATCG00340
|
PSAB |
-0.444 |
-0.022 |
| 517 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
-0.444 |
0.003 |
| 518 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.443 |
-0.772 |
| 519 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.442 |
-0.345 |
| 520 |
267165_at |
AT2G37710
|
LecRK-IV.1, L-type lectin receptor kinase IV.1, RLK, receptor lectin kinase |
-0.442 |
0.147 |
| 521 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.441 |
-0.513 |
| 522 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.440 |
-0.475 |
| 523 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.439 |
-0.105 |
| 524 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.439 |
-0.316 |
| 525 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.439 |
0.036 |
| 526 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.438 |
-0.060 |
| 527 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.437 |
0.009 |
| 528 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.436 |
-0.145 |
| 529 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.436 |
-0.040 |
| 530 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.435 |
0.928 |
| 531 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
-0.434 |
-0.217 |
| 532 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.433 |
0.081 |
| 533 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
-0.433 |
-0.027 |
| 534 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
-0.433 |
-0.179 |
| 535 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.432 |
-0.503 |
| 536 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.430 |
-0.145 |
| 537 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.430 |
-0.027 |
| 538 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.430 |
-0.101 |
| 539 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.429 |
-0.003 |
| 540 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.428 |
-0.006 |
| 541 |
247476_at |
AT5G62330
|
unknown |
-0.426 |
-0.068 |
| 542 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.426 |
-0.098 |
| 543 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.426 |
0.054 |
| 544 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.425 |
-0.349 |
| 545 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.423 |
-0.333 |
| 546 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.423 |
-0.559 |
| 547 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.423 |
0.020 |
| 548 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.423 |
-0.117 |
| 549 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.423 |
0.049 |
| 550 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.422 |
0.010 |
| 551 |
257154_at |
AT3G27210
|
unknown |
-0.422 |
0.793 |
| 552 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.422 |
-0.524 |
| 553 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.422 |
0.044 |
| 554 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
-0.421 |
-0.036 |
| 555 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.419 |
-0.020 |
| 556 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.419 |
0.916 |
| 557 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
-0.418 |
-0.062 |
| 558 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.418 |
-0.037 |
| 559 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.417 |
0.040 |
| 560 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.417 |
-0.204 |
| 561 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.417 |
-0.327 |
| 562 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.416 |
-0.183 |
| 563 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.416 |
0.133 |
| 564 |
252425_at |
AT3G47620
|
AtTCP14, TEOSINTE BRANCHED, cycloidea and PCF (TCP) 14, TCP14, TEOSINTE BRANCHED, cycloidea and PCF (TCP) 14 |
-0.416 |
0.135 |
| 565 |
259541_at |
AT1G20650
|
ASG5, ALTERED SEED GERMINATION 5 |
-0.415 |
-0.278 |
| 566 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.415 |
-0.014 |
| 567 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
-0.414 |
-0.300 |
| 568 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.414 |
-0.254 |
| 569 |
254816_at |
AT4G12440
|
APT4, adenine phosphoribosyl transferase 4 |
-0.412 |
0.023 |
| 570 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.412 |
-0.268 |
| 571 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.410 |
-0.076 |
| 572 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.409 |
0.793 |
| 573 |
252224_at |
AT3G49860
|
ARLA1B, ADP-ribosylation factor-like A1B, ATARLA1B, ADP-ribosylation factor-like A1B |
-0.408 |
-0.003 |
| 574 |
244961_at |
ATCG01040
|
YCF5 |
-0.408 |
-0.045 |
| 575 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.408 |
0.157 |
| 576 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.407 |
-0.074 |
| 577 |
246888_at |
AT5G26270
|
unknown |
-0.407 |
0.273 |
| 578 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.406 |
-0.374 |
| 579 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.406 |
0.021 |
| 580 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-0.406 |
0.027 |
| 581 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
-0.406 |
-0.061 |
| 582 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.406 |
0.544 |
| 583 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
-0.405 |
-0.188 |
| 584 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.404 |
-0.229 |
| 585 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.404 |
-0.305 |
| 586 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.404 |
0.016 |
| 587 |
254510_at |
AT4G20210
|
TPS08, terpene synthase 8 |
-0.404 |
0.019 |
| 588 |
256759_at |
AT3G25640
|
unknown |
-0.404 |
0.572 |
| 589 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.403 |
-0.543 |
| 590 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.402 |
-0.070 |
| 591 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.400 |
0.014 |
| 592 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.399 |
0.148 |
| 593 |
247357_at |
AT5G63710
|
[Leucine-rich repeat protein kinase family protein] |
-0.398 |
-0.219 |
| 594 |
251315_at |
AT3G61410
|
unknown |
-0.397 |
-0.044 |
| 595 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.397 |
-0.363 |
| 596 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.396 |
-0.296 |
| 597 |
249034_at |
AT5G44160
|
AtIDD8, IDD8, INDETERMINATE DOMAIN 8, NUC, nutcracker |
-0.396 |
-0.132 |
| 598 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.396 |
-0.255 |
| 599 |
252458_at |
AT3G47210
|
unknown |
-0.395 |
-0.031 |
| 600 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.394 |
0.060 |