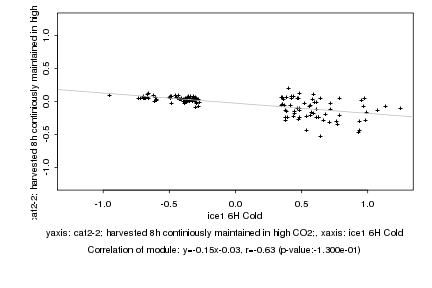
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ice1 6H Cold |
Log2 signal ratio
cat2-2; harvested 8h continiously maintained in high CO2; |
| 1 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.245 |
-0.105 |
| 2 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.127 |
-0.067 |
| 3 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.074 |
-0.122 |
| 4 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.989 |
-0.163 |
| 5 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.981 |
-0.283 |
| 6 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.973 |
0.058 |
| 7 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.963 |
-0.070 |
| 8 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.952 |
0.027 |
| 9 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.936 |
-0.427 |
| 10 |
264635_at |
AT1G65500
|
unknown |
0.936 |
-0.303 |
| 11 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.928 |
-0.456 |
| 12 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.784 |
0.051 |
| 13 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.782 |
-0.209 |
| 14 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.765 |
-0.335 |
| 15 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.757 |
-0.291 |
| 16 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.718 |
-0.028 |
| 17 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.718 |
-0.114 |
| 18 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.707 |
-0.314 |
| 19 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.678 |
-0.186 |
| 20 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.661 |
-0.287 |
| 21 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.640 |
-0.530 |
| 22 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.639 |
0.046 |
| 23 |
248889_at |
AT5G46230
|
unknown |
0.626 |
-0.241 |
| 24 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.625 |
-0.237 |
| 25 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.611 |
-0.012 |
| 26 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.608 |
-0.242 |
| 27 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.607 |
-0.120 |
| 28 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.592 |
-0.013 |
| 29 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.590 |
-0.169 |
| 30 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.588 |
0.110 |
| 31 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
0.577 |
0.037 |
| 32 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.572 |
-0.160 |
| 33 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.567 |
-0.055 |
| 34 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.564 |
-0.199 |
| 35 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.555 |
-0.075 |
| 36 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.533 |
-0.431 |
| 37 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.532 |
-0.216 |
| 38 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.518 |
-0.029 |
| 39 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.482 |
-0.134 |
| 40 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.481 |
-0.239 |
| 41 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.481 |
0.133 |
| 42 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.478 |
-0.099 |
| 43 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.475 |
0.056 |
| 44 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.475 |
-0.265 |
| 45 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.464 |
-0.102 |
| 46 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
0.463 |
0.059 |
| 47 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.446 |
-0.151 |
| 48 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.441 |
-0.169 |
| 49 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.434 |
-0.213 |
| 50 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.432 |
0.088 |
| 51 |
259514_at |
AT1G12480
|
CDI3, CARBON DIOXIDE INSENSITIVE 3, OZS1, OZONE-SENSITIVE 1, RCD3, RADICAL-INDUCED CELL DEATH 3, SLAC1, SLOW ANION CHANNEL-ASSOCIATED 1 |
0.423 |
0.053 |
| 52 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
0.418 |
0.082 |
| 53 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.416 |
-0.060 |
| 54 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.400 |
0.211 |
| 55 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.393 |
-0.230 |
| 56 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
0.386 |
-0.143 |
| 57 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.384 |
0.069 |
| 58 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.376 |
-0.230 |
| 59 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.375 |
-0.275 |
| 60 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.373 |
-0.132 |
| 61 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.364 |
-0.050 |
| 62 |
261401_at |
AT1G79640
|
[Protein kinase superfamily protein] |
0.363 |
0.038 |
| 63 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.355 |
0.063 |
| 64 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.354 |
-0.032 |
| 65 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.351 |
-0.040 |
| 66 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
0.345 |
-0.047 |
| 67 |
254494_at |
AT4G20050
|
QRT3, QUARTET 3 |
0.343 |
0.069 |
| 68 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.955 |
0.097 |
| 69 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.739 |
0.055 |
| 70 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.722 |
0.060 |
| 71 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.701 |
0.080 |
| 72 |
247878_at |
AT5G57760
|
unknown |
-0.698 |
0.061 |
| 73 |
252215_at |
AT3G50240
|
KICP-02 |
-0.696 |
0.062 |
| 74 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.688 |
0.051 |
| 75 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.685 |
0.059 |
| 76 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.669 |
0.072 |
| 77 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.665 |
0.111 |
| 78 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.664 |
0.125 |
| 79 |
250936_at |
AT5G03120
|
unknown |
-0.662 |
0.049 |
| 80 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.627 |
0.092 |
| 81 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.612 |
0.015 |
| 82 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.605 |
0.068 |
| 83 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.602 |
0.023 |
| 84 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.601 |
0.041 |
| 85 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.504 |
0.065 |
| 86 |
258367_at |
AT3G14370
|
WAG2 |
-0.494 |
0.084 |
| 87 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.493 |
0.052 |
| 88 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.491 |
0.085 |
| 89 |
260515_at |
AT1G51460
|
ABCG13, ATP-binding cassette G13 |
-0.484 |
-0.020 |
| 90 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.458 |
0.097 |
| 91 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.451 |
0.065 |
| 92 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.444 |
0.046 |
| 93 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.436 |
0.101 |
| 94 |
257769_at |
AT3G23050
|
AXR2, AUXIN RESISTANT 2, IAA7, indole-3-acetic acid 7 |
-0.428 |
0.033 |
| 95 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.426 |
0.042 |
| 96 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.416 |
0.039 |
| 97 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.414 |
0.038 |
| 98 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.410 |
0.058 |
| 99 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.398 |
0.007 |
| 100 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.395 |
0.044 |
| 101 |
255028_at |
AT4G09890
|
unknown |
-0.389 |
-0.020 |
| 102 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.387 |
0.039 |
| 103 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.383 |
0.005 |
| 104 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.383 |
0.029 |
| 105 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.382 |
0.052 |
| 106 |
250284_at |
AT5G13290
|
CRN, CORYNE, SOL2, SUPPRESSOR OF LLP1 2 |
-0.374 |
0.023 |
| 107 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
-0.373 |
-0.009 |
| 108 |
260696_at |
AT1G32520
|
unknown |
-0.372 |
0.027 |
| 109 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.370 |
0.075 |
| 110 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.363 |
0.022 |
| 111 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.358 |
0.087 |
| 112 |
251036_at |
AT5G02160
|
unknown |
-0.349 |
0.015 |
| 113 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.348 |
0.060 |
| 114 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.342 |
0.089 |
| 115 |
266335_at |
AT2G32440
|
ATKAO2, ARABIDOPSIS ENT-KAURENOIC ACID HYDROXYLASE 2, CYP88A4, KAO2, ent-kaurenoic acid hydroxylase 2 |
-0.336 |
0.028 |
| 116 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
-0.334 |
0.029 |
| 117 |
258204_at |
AT3G13960
|
AtGRF5, growth-regulating factor 5, GRF5, growth-regulating factor 5 |
-0.327 |
0.019 |
| 118 |
256035_at |
AT1G07140
|
SIRANBP |
-0.326 |
0.015 |
| 119 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.323 |
0.052 |
| 120 |
251701_at |
AT3G56650
|
PPD6, PsbP-domain protein 6 |
-0.323 |
0.021 |
| 121 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.322 |
0.074 |
| 122 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.319 |
0.024 |
| 123 |
248423_at |
AT5G51670
|
unknown |
-0.318 |
0.078 |
| 124 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.314 |
0.029 |
| 125 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.308 |
-0.087 |
| 126 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.308 |
0.075 |
| 127 |
261536_at |
AT1G01790
|
ATKEA1, K+ EFFLUX ANTIPORTER 1, KEA1, K+ efflux antiporter 1 |
-0.306 |
0.043 |
| 128 |
266481_at |
AT2G31070
|
TCP10, TCP domain protein 10 |
-0.303 |
0.029 |
| 129 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.300 |
0.060 |
| 130 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.297 |
-0.019 |
| 131 |
257026_at |
AT3G19200
|
unknown |
-0.296 |
-0.014 |
| 132 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.282 |
-0.062 |
| 133 |
266221_at |
AT2G28760
|
UXS6, UDP-XYL synthase 6 |
-0.281 |
0.036 |
| 134 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-0.272 |
-0.006 |