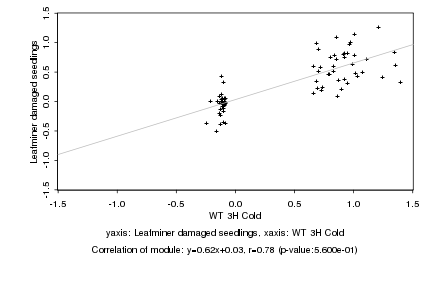
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT 3H Cold |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
1.393 |
0.333 |
| 2 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
1.349 |
0.615 |
| 3 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
1.345 |
0.837 |
| 4 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
1.237 |
0.422 |
| 5 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.209 |
1.263 |
| 6 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
1.103 |
0.716 |
| 7 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
1.070 |
0.507 |
| 8 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
1.030 |
0.432 |
| 9 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
1.014 |
0.489 |
| 10 |
253643_at |
AT4G29780
|
unknown |
1.006 |
0.791 |
| 11 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
1.002 |
1.153 |
| 12 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.984 |
0.635 |
| 13 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.965 |
1.010 |
| 14 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.960 |
0.976 |
| 15 |
248432_at |
AT5G51390
|
unknown |
0.946 |
0.318 |
| 16 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.940 |
0.828 |
| 17 |
256442_at |
AT3G10930
|
unknown |
0.922 |
0.757 |
| 18 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.921 |
0.826 |
| 19 |
264314_at |
AT1G70420
|
unknown |
0.917 |
0.387 |
| 20 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.909 |
0.806 |
| 21 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.893 |
0.216 |
| 22 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.864 |
0.365 |
| 23 |
248389_at |
AT5G51990
|
CBF4, C-repeat-binding factor 4, DREB1D, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1D |
0.861 |
0.090 |
| 24 |
253830_at |
AT4G27652
|
unknown |
0.849 |
0.727 |
| 25 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.847 |
1.091 |
| 26 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.831 |
0.788 |
| 27 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.823 |
0.511 |
| 28 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.823 |
0.606 |
| 29 |
262452_at |
AT1G11210
|
unknown |
0.797 |
0.762 |
| 30 |
257925_at |
AT3G23170
|
unknown |
0.789 |
0.469 |
| 31 |
255733_at |
AT1G25400
|
unknown |
0.785 |
0.475 |
| 32 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.731 |
0.254 |
| 33 |
250098_at |
AT5G17350
|
unknown |
0.721 |
0.193 |
| 34 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.712 |
0.581 |
| 35 |
246018_at |
AT5G10695
|
unknown |
0.699 |
0.515 |
| 36 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.695 |
0.900 |
| 37 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.687 |
0.225 |
| 38 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.680 |
0.998 |
| 39 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.678 |
0.345 |
| 40 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.660 |
0.607 |
| 41 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.655 |
0.146 |
| 42 |
253243_at |
AT4G34560
|
unknown |
-0.250 |
-0.359 |
| 43 |
255472_at |
AT4G02430
|
At-SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b, SR34b, Serine/Arginine-Rich Protein Splicing Factor 34b |
-0.215 |
0.009 |
| 44 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.163 |
-0.499 |
| 45 |
260277_at |
AT1G80520
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.158 |
0.004 |
| 46 |
265540_at |
AT2G15810
|
[Mutator-like transposase family, has a 1.5e-12 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.142 |
-0.031 |
| 47 |
266519_at |
AT2G35155
|
[Trypsin family protein] |
-0.137 |
-0.187 |
| 48 |
249841_at |
AT5G23520
|
[smr (Small MutS Related) domain-containing protein] |
-0.136 |
0.099 |
| 49 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.130 |
-0.227 |
| 50 |
253486_at |
AT4G31600
|
UTr7, UDP-galactose transporter 7 |
-0.128 |
-0.122 |
| 51 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
-0.128 |
-0.375 |
| 52 |
262193_at |
AT1G77840
|
[Translation initiation factor IF2/IF5] |
-0.122 |
0.047 |
| 53 |
258951_at |
AT3G01380
|
[transferases] |
-0.120 |
-0.030 |
| 54 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.120 |
0.432 |
| 55 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
-0.119 |
0.136 |
| 56 |
264542_at |
AT1G55830
|
unknown |
-0.118 |
0.003 |
| 57 |
263370_at |
AT2G20500
|
unknown |
-0.117 |
-0.032 |
| 58 |
253742_at |
AT4G28900
|
[copia-like retrotransposon family, has a 7.7e-236 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
-0.116 |
-0.074 |
| 59 |
252747_at |
AT3G43320
|
unknown |
-0.112 |
-0.011 |
| 60 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.111 |
-0.037 |
| 61 |
267249_at |
AT2G23040
|
unknown |
-0.111 |
0.002 |
| 62 |
265812_at |
AT2G18070
|
unknown |
-0.110 |
0.053 |
| 63 |
246285_at |
AT4G36980
|
unknown |
-0.106 |
-0.037 |
| 64 |
254246_at |
AT4G23250
|
CRK17, CYSTEINE-RICH RLK (RECEPTOR-LIKE PROTEIN KINASE) 17, DUF26-21, EMB1290, EMBRYO DEFECTIVE 1290, RKC1, RECEPTOR-LIKE KINASE THALIANACOL-0GENOMICLIBRARY(CLONTECH).THEPOSITIVEPHAGECLONES C-X8-C-X2-C CLASS 1 |
-0.105 |
-0.115 |
| 65 |
254168_at |
AT4G24250
|
ATMLO13, MILDEW RESISTANCE LOCUS O 13, MLO13, MILDEW RESISTANCE LOCUS O 13 |
-0.105 |
-0.023 |
| 66 |
267610_at |
AT2G26650
|
AKT1, K+ transporter 1, ATAKT1, KT1, K+ transporter 1 |
-0.103 |
0.339 |
| 67 |
252615_at |
AT3G45230
|
APAP1, ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN 1 |
-0.103 |
-0.349 |
| 68 |
250424_at |
AT5G10550
|
GTE2, global transcription factor group E2 |
-0.103 |
-0.163 |
| 69 |
246347_at |
AT3G56830
|
unknown |
-0.103 |
-0.073 |
| 70 |
248635_at |
AT5G49050
|
unknown |
-0.100 |
0.015 |
| 71 |
246264_at |
AT1G31814
|
FRL2, FRIGIDA like 2 |
-0.098 |
-0.035 |
| 72 |
246897_at |
AT5G25560
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.097 |
-0.038 |
| 73 |
246062_at |
AT5G19330
|
ARIA, ARM repeat protein interacting with ABF2 |
-0.097 |
-0.020 |
| 74 |
255666_at |
AT4G00390
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.096 |
0.041 |
| 75 |
262702_at |
AT1G16220
|
[Protein phosphatase 2C family protein] |
-0.095 |
-0.064 |
| 76 |
258917_at |
AT3G10630
|
[UDP-Glycosyltransferase superfamily protein] |
-0.095 |
-0.013 |
| 77 |
265300_at |
AT2G13950
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.093 |
0.066 |
| 78 |
250376_at |
AT5G11550
|
[ARM repeat superfamily protein] |
-0.092 |
-0.372 |
| 79 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.092 |
-0.033 |
| 80 |
256519_at |
AT1G66110
|
unknown |
-0.091 |
0.061 |
| 81 |
266169_at |
AT2G38900
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.091 |
-0.006 |