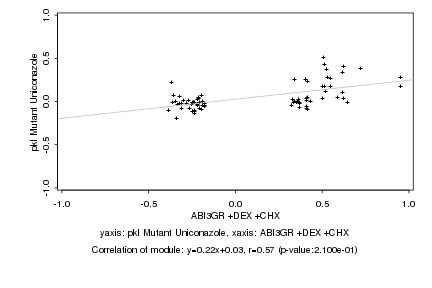
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABI3GR +DEX +CHX |
Log2 signal ratio
pkl Mutant Uniconazole |
| 1 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.949 |
0.282 |
| 2 |
251202_at |
AT3G63040
|
unknown |
0.947 |
0.179 |
| 3 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
0.717 |
0.391 |
| 4 |
266486_at |
AT2G47950
|
unknown |
0.641 |
-0.007 |
| 5 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.619 |
0.411 |
| 6 |
250118_at |
AT5G16460
|
[Putative adipose-regulatory protein (Seipin)] |
0.617 |
0.045 |
| 7 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.615 |
0.109 |
| 8 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.615 |
0.345 |
| 9 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
0.587 |
0.049 |
| 10 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
0.547 |
0.277 |
| 11 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.543 |
0.174 |
| 12 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.527 |
0.282 |
| 13 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.523 |
0.373 |
| 14 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
0.517 |
0.117 |
| 15 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.509 |
0.174 |
| 16 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.508 |
0.436 |
| 17 |
248915_at |
AT5G45690
|
unknown |
0.504 |
0.516 |
| 18 |
250010_at |
AT5G18450
|
[Integrase-type DNA-binding superfamily protein] |
0.499 |
0.174 |
| 19 |
266468_at |
AT2G47960
|
unknown |
0.496 |
0.037 |
| 20 |
246971_at |
AT5G24940
|
[Protein phosphatase 2C family protein] |
0.428 |
0.004 |
| 21 |
261170_at |
AT1G04910
|
[O-fucosyltransferase family protein] |
0.413 |
-0.086 |
| 22 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.410 |
0.048 |
| 23 |
262069_at |
AT1G80090
|
CBSX4, CBS domain containing protein 4 |
0.410 |
0.232 |
| 24 |
257056_at |
AT3G15350
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.407 |
-0.072 |
| 25 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
0.406 |
0.044 |
| 26 |
246548_at |
AT5G14910
|
[Heavy metal transport/detoxification superfamily protein ] |
0.406 |
0.014 |
| 27 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.404 |
-0.047 |
| 28 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.402 |
0.260 |
| 29 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.369 |
-0.007 |
| 30 |
259903_at |
AT1G74160
|
TRM4, TON1 Recruiting Motif 4 |
0.366 |
-0.063 |
| 31 |
267253_at |
AT2G22960
|
[alpha/beta-Hydrolases superfamily protein] |
0.364 |
-0.020 |
| 32 |
263458_at |
AT2G22290
|
ATRAB-H1D, ARABIDOPSIS RAB GTPASE HOMOLOG H1D, ATRAB6, ARABIDOPSIS RAB GTPASE HOMOLOG 6, ATRABH1D, RAB GTPase homolog H1D, RAB-H1D, RAB GTPASE HOMOLOG H1D, RABH1d, RAB GTPase homolog H1D |
0.362 |
0.033 |
| 33 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.353 |
0.007 |
| 34 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
0.347 |
-0.002 |
| 35 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.339 |
0.255 |
| 36 |
249330_at |
AT5G40970
|
unknown |
0.335 |
0.002 |
| 37 |
263858_at |
AT2G04370
|
unknown |
0.329 |
-0.006 |
| 38 |
253504_at |
AT4G31960
|
unknown |
0.326 |
0.031 |
| 39 |
253851_at |
AT4G28110
|
AtMYB41, myb domain protein 41, MYB41, myb domain protein 41 |
0.323 |
-0.044 |
| 40 |
246041_at |
AT5G19290
|
[alpha/beta-Hydrolases superfamily protein] |
-0.388 |
-0.098 |
| 41 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.371 |
0.231 |
| 42 |
266972_at |
AT2G39590
|
[Ribosomal protein S8 family protein] |
-0.368 |
-0.010 |
| 43 |
248181_at |
AT5G54290
|
CcdA |
-0.361 |
0.070 |
| 44 |
255932_at |
AT1G12720
|
[Mutator-like transposase family, has a 1.2e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.351 |
0.002 |
| 45 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.345 |
-0.190 |
| 46 |
248116_at |
AT5G55020
|
ATMYB120, MYB120, myb domain protein 120 |
-0.339 |
-0.030 |
| 47 |
260199_at |
AT1G67590
|
[Remorin family protein] |
-0.327 |
-0.015 |
| 48 |
253989_at |
AT4G26130
|
unknown |
-0.323 |
0.064 |
| 49 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
-0.316 |
-0.079 |
| 50 |
245279_at |
AT4G17270
|
[Mo25 family protein] |
-0.316 |
-0.018 |
| 51 |
263592_at |
AT2G01840
|
[non-LTR retrotransposon family (LINE), has a 9.6e-34 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
-0.300 |
0.014 |
| 52 |
257915_at |
AT3G23130
|
FLO10, FLORAL DEFECTIVE 10, FON1, FLORAL ORGAN NUMBER 1, SUP, SUPERMAN |
-0.285 |
-0.016 |
| 53 |
253800_at |
AT4G28160
|
[hydroxyproline-rich glycoprotein family protein] |
-0.276 |
0.015 |
| 54 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.269 |
-0.077 |
| 55 |
260210_at |
AT1G74420
|
ATFUT3, FUT3, fucosyltransferase 3 |
-0.258 |
-0.030 |
| 56 |
262818_at |
AT1G11755
|
LEW1, LEAF WILTING 1 |
-0.253 |
-0.007 |
| 57 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.251 |
-0.113 |
| 58 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.248 |
-0.114 |
| 59 |
257691_at |
AT3G12660
|
FLA14, FASCICLIN-like arabinogalactan protein 14 precursor |
-0.247 |
-0.005 |
| 60 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.241 |
-0.104 |
| 61 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.238 |
-0.103 |
| 62 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.238 |
-0.130 |
| 63 |
248052_at |
AT5G55800
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.237 |
-0.004 |
| 64 |
261545_at |
AT1G63530
|
unknown |
-0.223 |
-0.023 |
| 65 |
247554_at |
AT5G61010
|
ATEXO70E2, exocyst subunit exo70 family protein E2, EXO70E2, exocyst subunit exo70 family protein E2 |
-0.221 |
0.033 |
| 66 |
258478_at |
AT3G02710
|
[ARM repeat superfamily protein] |
-0.220 |
-0.045 |
| 67 |
253686_at |
AT4G29750
|
[CRS1 / YhbY (CRM) domain-containing protein] |
-0.213 |
0.043 |
| 68 |
260916_at |
AT1G02475
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.213 |
0.048 |
| 69 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.212 |
-0.080 |
| 70 |
248872_at |
AT5G46795
|
MSP2, microspore-specific promoter 2 |
-0.208 |
-0.011 |
| 71 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.201 |
0.075 |
| 72 |
253056_at |
AT4G37650
|
SGR7, SHOOT GRAVITROPISM 7, SHR, SHORT ROOT |
-0.200 |
-0.084 |
| 73 |
250927_at |
AT5G03270
|
LOG6, LONELY GUY 6 |
-0.195 |
-0.037 |
| 74 |
258506_at |
AT3G06520
|
[agenet domain-containing protein] |
-0.192 |
0.009 |
| 75 |
258165_at |
AT3G17900
|
unknown |
-0.183 |
-0.045 |
| 76 |
248375_at |
AT5G51710
|
ATKEA5, ARABIDOPSIS THALIANA K+ EFFLUX ANTIPORTER 5, KEA5, K+ efflux antiporter 5 |
-0.181 |
-0.047 |
| 77 |
251445_at |
AT3G59870
|
unknown |
-0.180 |
-0.021 |