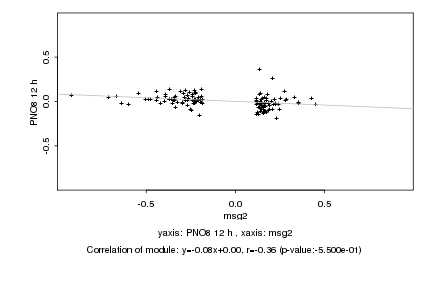
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msg2 |
Log2 signal ratio
PNO8 12 h |
| 1 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.446 |
-0.029 |
| 2 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.425 |
0.041 |
| 3 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.352 |
-0.012 |
| 4 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.349 |
-0.008 |
| 5 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.331 |
0.047 |
| 6 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.284 |
0.034 |
| 7 |
264481_at |
AT1G77200
|
[Integrase-type DNA-binding superfamily protein] |
0.275 |
0.022 |
| 8 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
0.272 |
0.121 |
| 9 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.252 |
0.044 |
| 10 |
260251_at |
AT1G74250
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.245 |
-0.089 |
| 11 |
247046_at |
AT5G66540
|
unknown |
0.236 |
-0.026 |
| 12 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.227 |
-0.181 |
| 13 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
0.222 |
-0.014 |
| 14 |
252898_at |
AT4G39500
|
CYP96A11, cytochrome P450, family 96, subfamily A, polypeptide 11 |
0.221 |
-0.033 |
| 15 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.216 |
0.031 |
| 16 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.209 |
-0.027 |
| 17 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.206 |
-0.080 |
| 18 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.205 |
0.262 |
| 19 |
257685_at |
AT3G12770
|
MEF22, mitochondrial editing factor 22 |
0.200 |
0.003 |
| 20 |
247257_at |
AT5G64760
|
RPN5B, regulatory particle non-ATPase subunit 5B |
0.190 |
-0.038 |
| 21 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.189 |
-0.083 |
| 22 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.185 |
-0.091 |
| 23 |
256927_at |
AT3G22550
|
unknown |
0.183 |
-0.022 |
| 24 |
245076_at |
AT2G23170
|
GH3.3 |
0.180 |
-0.012 |
| 25 |
255763_at |
AT1G16730
|
unknown |
0.177 |
0.085 |
| 26 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
0.173 |
0.039 |
| 27 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.172 |
0.012 |
| 28 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.170 |
-0.113 |
| 29 |
245293_at |
AT4G16660
|
[heat shock protein 70 (Hsp 70) family protein] |
0.168 |
-0.056 |
| 30 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.163 |
-0.109 |
| 31 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.162 |
0.055 |
| 32 |
246570_at |
AT5G14990
|
unknown |
0.162 |
-0.019 |
| 33 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
0.159 |
-0.004 |
| 34 |
251256_at |
AT3G62300
|
unknown |
0.159 |
-0.105 |
| 35 |
258585_at |
AT3G04340
|
emb2458, embryo defective 2458 |
0.158 |
-0.037 |
| 36 |
246334_at |
AT3G44850
|
[Protein kinase superfamily protein] |
0.158 |
-0.045 |
| 37 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
0.157 |
-0.065 |
| 38 |
263939_at |
AT2G36070
|
ATTIM44-2, translocase inner membrane subunit 44-2, TIM44-2, translocase inner membrane subunit 44-2 |
0.156 |
-0.124 |
| 39 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.156 |
-0.116 |
| 40 |
258840_at |
AT3G04620
|
DAN1, D NUCLDUO1-ACTIVATEEIC ACID BINDING PROTEIN 1 |
0.156 |
-0.081 |
| 41 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.153 |
-0.056 |
| 42 |
253807_at |
AT4G28280
|
LLG3, LORELEI-LIKE-GPI ANCHORED PROTEIN 3 |
0.150 |
0.039 |
| 43 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.146 |
-0.011 |
| 44 |
251012_at |
AT5G02580
|
unknown |
0.145 |
0.029 |
| 45 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.143 |
-0.081 |
| 46 |
248099_at |
AT5G55300
|
MGO1, MGOUN 1, TOP1, TOPOISOMERASE 1, TOP1ALPHA, DNA topoisomerase I alpha |
0.142 |
-0.038 |
| 47 |
259678_at |
AT1G77750
|
[Ribosomal protein S13/S18 family] |
0.141 |
-0.044 |
| 48 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.140 |
0.100 |
| 49 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.140 |
-0.017 |
| 50 |
255586_at |
AT4G01560
|
MEE49, maternal effect embryo arrest 49 |
0.139 |
-0.102 |
| 51 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.138 |
0.010 |
| 52 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
0.137 |
-0.018 |
| 53 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.136 |
-0.029 |
| 54 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.135 |
-0.076 |
| 55 |
251339_at |
AT3G60780
|
unknown |
0.133 |
0.082 |
| 56 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.130 |
0.361 |
| 57 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
0.130 |
-0.058 |
| 58 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.129 |
-0.135 |
| 59 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
0.127 |
-0.061 |
| 60 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.122 |
-0.114 |
| 61 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.121 |
-0.019 |
| 62 |
263488_at |
AT2G31840
|
MRL7-L, Mesophyll-cell RNAi Library line 7-like |
0.118 |
-0.029 |
| 63 |
254114_at |
AT4G24980
|
UMAMIT16-psi, Usually multiple acids move in and out Transporters 16-pseudogene |
0.118 |
0.036 |
| 64 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
0.118 |
0.018 |
| 65 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.118 |
0.011 |
| 66 |
247024_at |
AT5G66985
|
unknown |
0.117 |
-0.140 |
| 67 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.923 |
0.079 |
| 68 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.713 |
0.051 |
| 69 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.668 |
0.065 |
| 70 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.639 |
-0.015 |
| 71 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.603 |
-0.025 |
| 72 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.548 |
0.099 |
| 73 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.505 |
0.034 |
| 74 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.488 |
0.025 |
| 75 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.479 |
0.024 |
| 76 |
264774_at |
AT1G22890
|
unknown |
-0.447 |
0.117 |
| 77 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.444 |
0.018 |
| 78 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.439 |
0.052 |
| 79 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.422 |
-0.020 |
| 80 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.401 |
0.006 |
| 81 |
252356_at |
AT3G48370
|
[copia-like retrotransposon family, has a 7.2e-35 P-value blast match to GB:CAA72990 open reading frame 2 (Ty1_Copia-element) (Brassica oleracea)] |
-0.396 |
0.084 |
| 82 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.394 |
0.066 |
| 83 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.375 |
0.146 |
| 84 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.373 |
0.033 |
| 85 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
-0.370 |
0.032 |
| 86 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.360 |
0.025 |
| 87 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.356 |
-0.018 |
| 88 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.352 |
0.019 |
| 89 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.345 |
0.055 |
| 90 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.345 |
0.032 |
| 91 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.342 |
0.019 |
| 92 |
256602_at |
AT3G28310
|
unknown |
-0.339 |
-0.065 |
| 93 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.337 |
0.063 |
| 94 |
258490_at |
AT3G02670
|
[Glycine-rich protein family] |
-0.329 |
-0.008 |
| 95 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.308 |
0.116 |
| 96 |
266877_at |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
-0.302 |
-0.001 |
| 97 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.300 |
-0.018 |
| 98 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.296 |
0.091 |
| 99 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.291 |
0.047 |
| 100 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.284 |
0.127 |
| 101 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.282 |
0.013 |
| 102 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.270 |
0.014 |
| 103 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.270 |
-0.037 |
| 104 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.270 |
0.069 |
| 105 |
251727_at |
AT3G56290
|
unknown |
-0.268 |
0.042 |
| 106 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.258 |
0.109 |
| 107 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.257 |
-0.085 |
| 108 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.249 |
-0.095 |
| 109 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.246 |
0.090 |
| 110 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
-0.242 |
0.059 |
| 111 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.242 |
0.022 |
| 112 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
-0.240 |
-0.018 |
| 113 |
252765_at |
AT3G42800
|
unknown |
-0.234 |
0.022 |
| 114 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.234 |
-0.014 |
| 115 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.230 |
0.028 |
| 116 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.230 |
0.129 |
| 117 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.229 |
0.109 |
| 118 |
262421_at |
AT1G50290
|
unknown |
-0.228 |
0.100 |
| 119 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.227 |
0.009 |
| 120 |
249010_at |
AT5G44580
|
unknown |
-0.224 |
0.033 |
| 121 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.221 |
0.011 |
| 122 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.218 |
0.014 |
| 123 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.209 |
0.055 |
| 124 |
267119_at |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
-0.209 |
0.013 |
| 125 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.202 |
-0.151 |
| 126 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.201 |
-0.001 |
| 127 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.196 |
0.034 |
| 128 |
249622_at |
AT5G37550
|
unknown |
-0.195 |
-0.007 |
| 129 |
250128_at |
AT5G16540
|
ZFN3, zinc finger nuclease 3 |
-0.194 |
0.065 |
| 130 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.194 |
0.141 |
| 131 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.189 |
-0.017 |
| 132 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.189 |
0.032 |