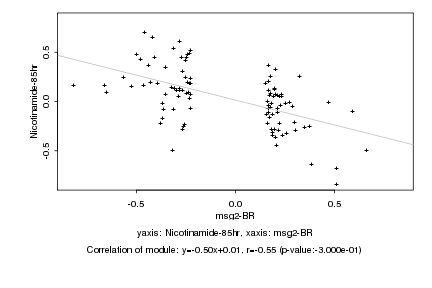
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msg2-BR |
Log2 signal ratio
Nicotinamide-85hr |
| 1 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.658 |
-0.488 |
| 2 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.588 |
-0.097 |
| 3 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.507 |
-0.678 |
| 4 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.507 |
-0.832 |
| 5 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.470 |
-0.005 |
| 6 |
252888_at |
AT4G39210
|
APL3 |
0.382 |
-0.630 |
| 7 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.372 |
-0.253 |
| 8 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.347 |
-0.255 |
| 9 |
263377_at |
AT2G40220
|
ABI4, ABA INSENSITIVE 4, ATABI4, GIN6, GLUCOSE INSENSITIVE 6, ISI3, IMPAIRED SUCROSE INDUCTION 3, SAN5, SALOBRENO 5, SIS5, SUGAR-INSENSITIVE 5, SUN6, SUCROSE UNCOUPLED 6 |
0.323 |
0.260 |
| 10 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.302 |
-0.287 |
| 11 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.296 |
-0.212 |
| 12 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.284 |
-0.049 |
| 13 |
247070_at |
AT5G66815
|
unknown |
0.271 |
-0.005 |
| 14 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.255 |
-0.321 |
| 15 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
0.251 |
-0.017 |
| 16 |
265327_at |
AT2G18210
|
unknown |
0.236 |
-0.336 |
| 17 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.232 |
0.053 |
| 18 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.229 |
0.074 |
| 19 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.227 |
-0.032 |
| 20 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
0.222 |
-0.220 |
| 21 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
0.219 |
0.053 |
| 22 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.217 |
-0.286 |
| 23 |
247190_at |
AT5G65420
|
CYCD4;1, CYCLIN D4;1 |
0.212 |
-0.104 |
| 24 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.211 |
0.065 |
| 25 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.211 |
-0.068 |
| 26 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.203 |
-0.445 |
| 27 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.201 |
0.334 |
| 28 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.200 |
-0.358 |
| 29 |
254009_at |
AT4G26390
|
[Pyruvate kinase family protein] |
0.200 |
0.076 |
| 30 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.197 |
-0.276 |
| 31 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.196 |
0.141 |
| 32 |
266049_at |
AT2G40780
|
[Nucleic acid-binding, OB-fold-like protein] |
0.195 |
0.128 |
| 33 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.188 |
0.059 |
| 34 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
0.183 |
-0.308 |
| 35 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
0.183 |
-0.128 |
| 36 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.183 |
-0.344 |
| 37 |
258966_at |
AT3G10690
|
GYRA, DNA GYRASE A |
0.179 |
-0.010 |
| 38 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.179 |
-0.276 |
| 39 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.176 |
0.263 |
| 40 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.175 |
-0.058 |
| 41 |
248211_at |
AT5G54010
|
[UDP-Glycosyltransferase superfamily protein] |
0.173 |
0.083 |
| 42 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.171 |
0.067 |
| 43 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.168 |
-0.161 |
| 44 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.166 |
0.210 |
| 45 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.165 |
-0.040 |
| 46 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.164 |
0.370 |
| 47 |
246210_at |
AT4G36420
|
[Ribosomal protein L12 family protein] |
0.164 |
-0.105 |
| 48 |
265863_at |
AT2G01770
|
ATVIT1, VIT1, vacuolar iron transporter 1 |
0.163 |
0.112 |
| 49 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
0.162 |
-0.070 |
| 50 |
256927_at |
AT3G22550
|
unknown |
0.159 |
0.002 |
| 51 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
0.158 |
-0.221 |
| 52 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.153 |
-0.129 |
| 53 |
247178_at |
AT5G65205
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.149 |
0.183 |
| 54 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.823 |
0.172 |
| 55 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.664 |
0.163 |
| 56 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
-0.654 |
0.101 |
| 57 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.568 |
0.245 |
| 58 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
-0.528 |
0.154 |
| 59 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.504 |
0.487 |
| 60 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.484 |
0.433 |
| 61 |
246684_at |
AT5G33340
|
CDR1, CONSTITUTIVE DISEASE RESISTANCE 1 |
-0.466 |
0.164 |
| 62 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.463 |
0.707 |
| 63 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.443 |
0.369 |
| 64 |
256097_at |
AT1G13670
|
unknown |
-0.432 |
0.203 |
| 65 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.422 |
0.655 |
| 66 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.413 |
0.451 |
| 67 |
266392_at |
AT2G41280
|
ATM10, M10 |
-0.394 |
0.186 |
| 68 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.381 |
-0.217 |
| 69 |
263556_at |
AT2G16365
|
[F-box family protein] |
-0.370 |
-0.169 |
| 70 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.370 |
-0.017 |
| 71 |
263892_at |
AT2G36890
|
ATMYB38, MYB DOMAIN PROTEIN 38, BIT1, BLUE INSENSITIVE TRAIT 1, MYB38, MYB DOMAIN PROTEIN 38, RAX2, REGULATOR OF AXILLARY MERISTEMS 2 |
-0.367 |
-0.078 |
| 72 |
265590_at |
AT2G20160
|
ASK17, ARABIDOPSIS SKP1-LIKE 17, MEO, MEIDOS |
-0.358 |
0.080 |
| 73 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.354 |
0.352 |
| 74 |
266611_at |
AT2G14960
|
GH3.1 |
-0.328 |
0.147 |
| 75 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.321 |
-0.490 |
| 76 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.316 |
-0.073 |
| 77 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.315 |
0.548 |
| 78 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.309 |
0.136 |
| 79 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.309 |
0.137 |
| 80 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.302 |
0.120 |
| 81 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.292 |
0.058 |
| 82 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.287 |
0.142 |
| 83 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.283 |
0.617 |
| 84 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.283 |
0.114 |
| 85 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.275 |
0.447 |
| 86 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.272 |
-0.283 |
| 87 |
266877_at |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
-0.270 |
0.113 |
| 88 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.268 |
0.309 |
| 89 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.263 |
-0.250 |
| 90 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.260 |
-0.223 |
| 91 |
258468_at |
AT3G06070
|
unknown |
-0.254 |
0.249 |
| 92 |
260004_at |
AT1G67860
|
unknown |
-0.253 |
0.418 |
| 93 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.250 |
0.089 |
| 94 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.249 |
0.451 |
| 95 |
253832_at |
AT4G27654
|
unknown |
-0.247 |
0.484 |
| 96 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.244 |
0.194 |
| 97 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.238 |
0.094 |
| 98 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.237 |
0.035 |
| 99 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.237 |
0.490 |
| 100 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-0.237 |
0.183 |
| 101 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.231 |
0.527 |
| 102 |
266097_at |
AT2G37970
|
AtHBP2, HBP2, haem-binding protein 2, SOUL-1 |
-0.231 |
0.241 |
| 103 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
-0.229 |
0.080 |
| 104 |
264565_at |
AT1G05280
|
unknown |
-0.228 |
-0.066 |
| 105 |
250023_at |
AT5G18220
|
[O-Glycosyl hydrolases family 17 protein] |
-0.228 |
0.189 |