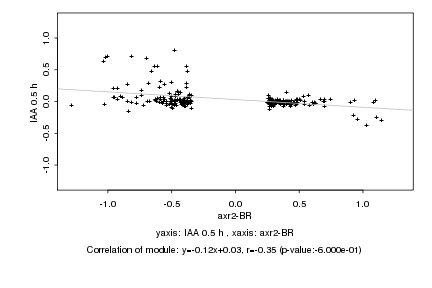
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
axr2-BR |
Log2 signal ratio
IAA 0.5 h |
| 1 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.143 |
-0.290 |
| 2 |
261619_x_at |
AT1G33130
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-125 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.102 |
-0.245 |
| 3 |
260861_at |
AT1G43840
|
[CACTA-like transposase family (Ptta/En/Spm), has a 5.2e-121 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.098 |
0.023 |
| 4 |
263479_x_at |
AT2G04000
|
unknown |
1.081 |
-0.002 |
| 5 |
263675_x_at |
AT2G04770
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.0e-20 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.020 |
-0.362 |
| 6 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.956 |
-0.275 |
| 7 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.929 |
0.021 |
| 8 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.924 |
-0.206 |
| 9 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.895 |
-0.003 |
| 10 |
261186_at |
AT1G34530
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.8e-116 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.741 |
0.037 |
| 11 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.696 |
-0.073 |
| 12 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
0.695 |
0.047 |
| 13 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.695 |
0.004 |
| 14 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.685 |
0.008 |
| 15 |
255805_at |
AT4G10240
|
bbx23, B-box domain protein 23 |
0.661 |
0.036 |
| 16 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.627 |
-0.016 |
| 17 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.622 |
-0.020 |
| 18 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.616 |
-0.009 |
| 19 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.610 |
-0.034 |
| 20 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
0.599 |
-0.003 |
| 21 |
249259_at |
AT5G41660
|
unknown |
0.579 |
-0.062 |
| 22 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.568 |
0.104 |
| 23 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.539 |
0.011 |
| 24 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.535 |
-0.038 |
| 25 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
0.533 |
0.088 |
| 26 |
263338_at |
AT2G05000
|
unknown |
0.503 |
0.018 |
| 27 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.498 |
0.037 |
| 28 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.485 |
-0.014 |
| 29 |
266738_at |
AT2G47010
|
unknown |
0.481 |
0.046 |
| 30 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.473 |
0.019 |
| 31 |
261180_at |
AT1G34590
|
unknown |
0.473 |
0.040 |
| 32 |
254522_at |
AT4G19980
|
unknown |
0.473 |
-0.032 |
| 33 |
255763_at |
AT1G16730
|
unknown |
0.471 |
-0.024 |
| 34 |
256211_at |
AT1G50960
|
ATGA2OX7, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 7, GA2OX7, gibberellin 2-oxidase 7 |
0.468 |
-0.055 |
| 35 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.462 |
-0.013 |
| 36 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.455 |
-0.007 |
| 37 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.451 |
-0.014 |
| 38 |
252888_at |
AT4G39210
|
APL3 |
0.437 |
-0.045 |
| 39 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.436 |
-0.024 |
| 40 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.433 |
0.023 |
| 41 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.427 |
0.000 |
| 42 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.426 |
-0.021 |
| 43 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.424 |
-0.076 |
| 44 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.420 |
0.009 |
| 45 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.416 |
-0.037 |
| 46 |
246552_at |
AT5G15420
|
unknown |
0.415 |
-0.017 |
| 47 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.414 |
0.001 |
| 48 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
0.407 |
0.001 |
| 49 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.406 |
-0.012 |
| 50 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.403 |
-0.031 |
| 51 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.399 |
0.155 |
| 52 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.397 |
0.005 |
| 53 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.396 |
-0.035 |
| 54 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.385 |
0.005 |
| 55 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.381 |
-0.046 |
| 56 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.378 |
-0.007 |
| 57 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.376 |
0.025 |
| 58 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
0.374 |
0.014 |
| 59 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
0.374 |
-0.076 |
| 60 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
0.370 |
-0.018 |
| 61 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.369 |
0.028 |
| 62 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.369 |
-0.001 |
| 63 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.362 |
-0.020 |
| 64 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.357 |
-0.022 |
| 65 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
0.351 |
-0.021 |
| 66 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.351 |
0.009 |
| 67 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
0.350 |
-0.001 |
| 68 |
264343_at |
AT1G11850
|
unknown |
0.349 |
0.023 |
| 69 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.348 |
-0.041 |
| 70 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.346 |
0.016 |
| 71 |
246268_at |
AT1G31800
|
CYP97A3, cytochrome P450, family 97, subfamily A, polypeptide 3, LUT5, LUTEIN DEFICIENT 5 |
0.344 |
-0.008 |
| 72 |
260877_at |
AT1G21500
|
unknown |
0.338 |
0.018 |
| 73 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.333 |
-0.005 |
| 74 |
259363_at |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
0.333 |
-0.013 |
| 75 |
257334_at |
ATMG01370
|
ORF111D |
0.332 |
-0.003 |
| 76 |
245007_at |
ATCG00350
|
PSAA |
0.331 |
0.003 |
| 77 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.329 |
0.010 |
| 78 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.328 |
0.001 |
| 79 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.323 |
0.042 |
| 80 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.318 |
-0.002 |
| 81 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.318 |
0.008 |
| 82 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.318 |
0.013 |
| 83 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.316 |
-0.019 |
| 84 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.312 |
0.031 |
| 85 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.312 |
-0.021 |
| 86 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.310 |
-0.025 |
| 87 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.304 |
-0.013 |
| 88 |
244998_at |
ATCG00180
|
RPOC1 |
0.301 |
-0.021 |
| 89 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.298 |
-0.006 |
| 90 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.298 |
-0.022 |
| 91 |
244967_at |
ATCG00630
|
PSAJ |
0.296 |
-0.000 |
| 92 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
0.296 |
-0.030 |
| 93 |
251012_at |
AT5G02580
|
unknown |
0.295 |
-0.059 |
| 94 |
254009_at |
AT4G26390
|
[Pyruvate kinase family protein] |
0.292 |
0.026 |
| 95 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.292 |
-0.076 |
| 96 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
0.291 |
0.002 |
| 97 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.290 |
-0.001 |
| 98 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.290 |
0.036 |
| 99 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
0.284 |
0.006 |
| 100 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.284 |
-0.029 |
| 101 |
255447_at |
AT4G02790
|
EMB3129, EMBRYO DEFECTIVE 3129 |
0.283 |
-0.005 |
| 102 |
249704_at |
AT5G35550
|
ATMYB123, MYB DOMAIN PROTEIN 123, ATTT2, MYB123, MYB DOMAIN PROTEIN 123, TT2, TRANSPARENT TESTA 2 |
0.280 |
0.037 |
| 103 |
265819_at |
AT2G17972
|
unknown |
0.278 |
0.007 |
| 104 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.278 |
0.010 |
| 105 |
257317_at |
ATMG01060
|
ORF107G |
0.278 |
0.017 |
| 106 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.277 |
-0.063 |
| 107 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.275 |
0.018 |
| 108 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.274 |
0.062 |
| 109 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.273 |
0.011 |
| 110 |
256927_at |
AT3G22550
|
unknown |
0.270 |
-0.044 |
| 111 |
253525_at |
AT4G31330
|
unknown |
0.269 |
-0.041 |
| 112 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.269 |
-0.014 |
| 113 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.268 |
0.006 |
| 114 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.267 |
-0.018 |
| 115 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.267 |
-0.002 |
| 116 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.264 |
-0.074 |
| 117 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.264 |
0.015 |
| 118 |
258966_at |
AT3G10690
|
GYRA, DNA GYRASE A |
0.264 |
-0.025 |
| 119 |
267580_at |
AT2G41990
|
unknown |
0.262 |
0.009 |
| 120 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.260 |
-0.114 |
| 121 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.260 |
-0.034 |
| 122 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.260 |
0.061 |
| 123 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.259 |
-0.004 |
| 124 |
264133_at |
AT1G79080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.258 |
-0.011 |
| 125 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.257 |
0.051 |
| 126 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.255 |
0.102 |
| 127 |
267630_at |
AT2G42130
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.254 |
-0.014 |
| 128 |
260264_at |
AT1G68500
|
unknown |
0.253 |
-0.031 |
| 129 |
251032_at |
AT5G02030
|
BLH9, BEL1-LIKE HOMEODOMAIN 9, BLR, BELLRINGER, HB-6, LSN, LARSON, PNY, PENNYWISE, RPL, REPLUMLESS, VAN, VAAMANA |
0.252 |
0.051 |
| 130 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.251 |
-0.001 |
| 131 |
251958_at |
AT3G53560
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.251 |
-0.005 |
| 132 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-1.291 |
-0.049 |
| 133 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-1.042 |
0.638 |
| 134 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-1.034 |
-0.045 |
| 135 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-1.025 |
0.694 |
| 136 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-1.004 |
0.716 |
| 137 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.963 |
0.071 |
| 138 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.959 |
0.215 |
| 139 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.956 |
0.064 |
| 140 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.932 |
0.212 |
| 141 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.930 |
0.047 |
| 142 |
264341_at |
AT1G70270
|
unknown |
-0.906 |
0.087 |
| 143 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.893 |
0.065 |
| 144 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-0.850 |
0.002 |
| 145 |
255028_at |
AT4G09890
|
unknown |
-0.848 |
0.268 |
| 146 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.839 |
-0.156 |
| 147 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.817 |
0.712 |
| 148 |
246684_at |
AT5G33340
|
CDR1, CONSTITUTIVE DISEASE RESISTANCE 1 |
-0.816 |
-0.013 |
| 149 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.779 |
-0.026 |
| 150 |
266392_at |
AT2G41280
|
ATM10, M10 |
-0.778 |
0.079 |
| 151 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.740 |
0.105 |
| 152 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.738 |
0.185 |
| 153 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.727 |
-0.054 |
| 154 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.703 |
0.679 |
| 155 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.690 |
0.011 |
| 156 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.684 |
0.291 |
| 157 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.675 |
0.009 |
| 158 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.664 |
0.476 |
| 159 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.638 |
0.567 |
| 160 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.633 |
0.036 |
| 161 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
-0.633 |
0.030 |
| 162 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.628 |
0.035 |
| 163 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.623 |
0.011 |
| 164 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.617 |
0.063 |
| 165 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.613 |
0.555 |
| 166 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.600 |
-0.005 |
| 167 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.597 |
0.233 |
| 168 |
251751_at |
AT3G55720
|
unknown |
-0.594 |
0.064 |
| 169 |
255957_at |
AT1G22160
|
unknown |
-0.593 |
0.032 |
| 170 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.588 |
0.324 |
| 171 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
-0.583 |
-0.006 |
| 172 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.573 |
-0.017 |
| 173 |
261772_at |
AT1G76240
|
unknown |
-0.569 |
0.073 |
| 174 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.567 |
0.003 |
| 175 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.562 |
0.270 |
| 176 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.559 |
0.043 |
| 177 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.547 |
0.004 |
| 178 |
265590_at |
AT2G20160
|
ASK17, ARABIDOPSIS SKP1-LIKE 17, MEO, MEIDOS |
-0.544 |
-0.023 |
| 179 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.543 |
-0.061 |
| 180 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.520 |
-0.041 |
| 181 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.520 |
0.137 |
| 182 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.515 |
0.008 |
| 183 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.511 |
0.060 |
| 184 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.508 |
0.090 |
| 185 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.507 |
0.035 |
| 186 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.506 |
-0.029 |
| 187 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
-0.505 |
-0.030 |
| 188 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.505 |
0.012 |
| 189 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.502 |
0.310 |
| 190 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.502 |
-0.089 |
| 191 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.501 |
0.027 |
| 192 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.500 |
-0.033 |
| 193 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
-0.494 |
-0.101 |
| 194 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-0.485 |
-0.051 |
| 195 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.483 |
0.012 |
| 196 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.482 |
0.019 |
| 197 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.479 |
0.819 |
| 198 |
249622_at |
AT5G37550
|
unknown |
-0.477 |
0.090 |
| 199 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.475 |
0.003 |
| 200 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.475 |
0.120 |
| 201 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
-0.473 |
0.029 |
| 202 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.470 |
-0.030 |
| 203 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.468 |
-0.048 |
| 204 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.461 |
0.163 |
| 205 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
-0.460 |
0.018 |
| 206 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.459 |
0.164 |
| 207 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.456 |
0.038 |
| 208 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.452 |
0.156 |
| 209 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.447 |
0.114 |
| 210 |
267085_at |
AT2G32620
|
ATCSLB02, cellulose synthase-like B, ATCSLB2, CELLULOSE SYNTHASE LIKE B2, CSLB02, CELLULOSE SYNTHASE LIKE B2, CSLB02, cellulose synthase-like B |
-0.446 |
0.027 |
| 211 |
253859_at |
AT4G27657
|
unknown |
-0.446 |
0.030 |
| 212 |
248674_at |
AT5G48800
|
[Phototropic-responsive NPH3 family protein] |
-0.443 |
0.033 |
| 213 |
252103_at |
AT3G51410
|
unknown |
-0.438 |
0.143 |
| 214 |
258468_at |
AT3G06070
|
unknown |
-0.437 |
-0.007 |
| 215 |
262085_at |
AT1G56060
|
unknown |
-0.437 |
0.046 |
| 216 |
266548_at |
AT2G35210
|
AGD10, MEE28, MATERNAL EFFECT EMBRYO ARREST 28, RPA, root and pollen arfgap |
-0.435 |
0.022 |
| 217 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
-0.430 |
0.010 |
| 218 |
260178_at |
AT1G70720
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.430 |
-0.008 |
| 219 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
-0.423 |
-0.032 |
| 220 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.421 |
-0.015 |
| 221 |
266877_at |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
-0.420 |
0.014 |
| 222 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.418 |
-0.005 |
| 223 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.415 |
0.043 |
| 224 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.411 |
-0.010 |
| 225 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.409 |
-0.002 |
| 226 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.408 |
-0.057 |
| 227 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.407 |
0.059 |
| 228 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.407 |
-0.024 |
| 229 |
257295_at |
AT3G17420
|
GPK1, glyoxysomal protein kinase 1 |
-0.406 |
0.030 |
| 230 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.406 |
-0.068 |
| 231 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.405 |
0.021 |
| 232 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.405 |
-0.015 |
| 233 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
-0.404 |
0.025 |
| 234 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.404 |
-0.021 |
| 235 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.403 |
-0.069 |
| 236 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
-0.402 |
0.018 |
| 237 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.399 |
-0.078 |
| 238 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.396 |
-0.008 |
| 239 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.389 |
0.221 |
| 240 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.389 |
0.010 |
| 241 |
263002_at |
AT1G54200
|
unknown |
-0.386 |
0.294 |
| 242 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.385 |
0.553 |
| 243 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.383 |
-0.042 |
| 244 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.381 |
0.017 |
| 245 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.380 |
0.478 |
| 246 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.378 |
0.071 |
| 247 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
-0.376 |
0.010 |
| 248 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.375 |
-0.012 |
| 249 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.373 |
-0.021 |
| 250 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.371 |
0.079 |
| 251 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.365 |
0.068 |
| 252 |
246063_at |
AT5G19340
|
unknown |
-0.364 |
0.122 |
| 253 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.364 |
-0.005 |
| 254 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.364 |
0.109 |
| 255 |
264314_at |
AT1G70420
|
unknown |
-0.358 |
0.027 |
| 256 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.357 |
0.006 |
| 257 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.354 |
0.031 |
| 258 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.353 |
-0.016 |
| 259 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.352 |
-0.099 |
| 260 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.350 |
0.100 |
| 261 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.350 |
0.005 |