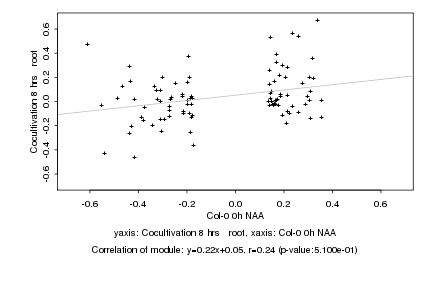
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 0h NAA |
Log2 signal ratio
Cocultivation 8 hrs root |
| 1 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.353 |
0.010 |
| 2 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.351 |
-0.125 |
| 3 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.338 |
0.679 |
| 4 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.320 |
0.196 |
| 5 |
263475_at |
AT2G31945
|
unknown |
0.316 |
0.363 |
| 6 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.308 |
0.088 |
| 7 |
258878_at |
AT3G03170
|
unknown |
0.308 |
-0.134 |
| 8 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.304 |
0.009 |
| 9 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.303 |
0.199 |
| 10 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.294 |
0.047 |
| 11 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
0.286 |
-0.018 |
| 12 |
249941_at |
AT5G22270
|
unknown |
0.273 |
0.157 |
| 13 |
248464_at |
AT5G51160
|
[Ankyrin repeat family protein] |
0.258 |
-0.084 |
| 14 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.257 |
0.540 |
| 15 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.234 |
0.570 |
| 16 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.233 |
-0.041 |
| 17 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.220 |
-0.098 |
| 18 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.214 |
0.053 |
| 19 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.212 |
0.284 |
| 20 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.211 |
-0.080 |
| 21 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.210 |
-0.179 |
| 22 |
263931_at |
AT2G36220
|
unknown |
0.204 |
0.205 |
| 23 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.194 |
-0.109 |
| 24 |
266835_at |
AT2G29990
|
NDA2, alternative NAD(P)H dehydrogenase 2 |
0.194 |
0.303 |
| 25 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.185 |
0.061 |
| 26 |
263757_at |
AT2G21430
|
[Papain family cysteine protease] |
0.182 |
0.044 |
| 27 |
259340_at |
AT3G03870
|
unknown |
0.180 |
0.219 |
| 28 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.176 |
-0.030 |
| 29 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.172 |
0.023 |
| 30 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.168 |
0.394 |
| 31 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.166 |
0.325 |
| 32 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
0.166 |
0.013 |
| 33 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.164 |
-0.017 |
| 34 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
0.163 |
0.013 |
| 35 |
260875_at |
AT1G21410
|
SKP2A |
0.161 |
0.169 |
| 36 |
260909_at |
AT1G02670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.161 |
-0.009 |
| 37 |
263370_at |
AT2G20500
|
unknown |
0.158 |
-0.024 |
| 38 |
250389_at |
AT5G11320
|
AtYUC4, YUC4, YUCCA4 |
0.153 |
-0.032 |
| 39 |
261641_at |
AT1G27670
|
unknown |
0.152 |
-0.022 |
| 40 |
267554_at |
AT2G32790
|
[Ubiquitin-conjugating enzyme family protein] |
0.146 |
0.087 |
| 41 |
248505_at |
AT5G50360
|
unknown |
0.145 |
0.001 |
| 42 |
248706_at |
AT5G48530
|
unknown |
0.145 |
0.004 |
| 43 |
260044_at |
AT1G73655
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.143 |
0.030 |
| 44 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.142 |
0.533 |
| 45 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
0.141 |
0.067 |
| 46 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.139 |
0.263 |
| 47 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
0.138 |
0.148 |
| 48 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
0.138 |
-0.032 |
| 49 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
0.134 |
0.007 |
| 50 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.613 |
0.480 |
| 51 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.554 |
-0.026 |
| 52 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.543 |
-0.424 |
| 53 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.488 |
0.032 |
| 54 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.467 |
0.126 |
| 55 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.438 |
0.293 |
| 56 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.438 |
-0.259 |
| 57 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.434 |
0.170 |
| 58 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.432 |
-0.199 |
| 59 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.420 |
-0.458 |
| 60 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.420 |
0.021 |
| 61 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
-0.389 |
-0.126 |
| 62 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.383 |
-0.151 |
| 63 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.375 |
-0.045 |
| 64 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.344 |
-0.197 |
| 65 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.337 |
0.127 |
| 66 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.328 |
0.095 |
| 67 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.323 |
0.018 |
| 68 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.313 |
0.004 |
| 69 |
260656_at |
AT1G19380
|
unknown |
-0.311 |
0.099 |
| 70 |
251335_at |
AT3G61400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.310 |
-0.141 |
| 71 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.308 |
-0.243 |
| 72 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
-0.301 |
0.200 |
| 73 |
263436_at |
AT2G28690
|
unknown |
-0.293 |
-0.147 |
| 74 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.276 |
-0.120 |
| 75 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.275 |
-0.073 |
| 76 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.272 |
-0.041 |
| 77 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.269 |
0.018 |
| 78 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.266 |
0.039 |
| 79 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.248 |
0.153 |
| 80 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.222 |
0.063 |
| 81 |
252205_at |
AT3G50350
|
unknown |
-0.220 |
0.043 |
| 82 |
249561_at |
AT5G38340
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.217 |
-0.097 |
| 83 |
245886_at |
AT5G09510
|
[Ribosomal protein S19 family protein] |
-0.216 |
-0.080 |
| 84 |
246839_at |
AT5G26720
|
unknown |
-0.200 |
0.012 |
| 85 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.198 |
-0.022 |
| 86 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.198 |
0.164 |
| 87 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.195 |
0.373 |
| 88 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.192 |
0.204 |
| 89 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.191 |
-0.092 |
| 90 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.188 |
0.033 |
| 91 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
-0.186 |
-0.252 |
| 92 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
-0.185 |
-0.022 |
| 93 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.183 |
0.049 |
| 94 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
-0.182 |
-0.125 |
| 95 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.180 |
-0.115 |
| 96 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.178 |
0.029 |
| 97 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.177 |
-0.361 |