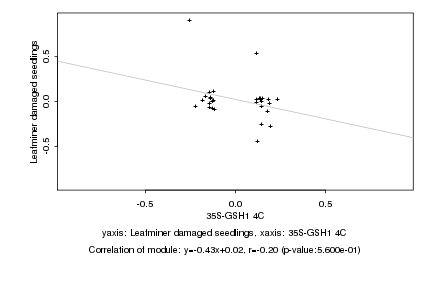
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S-GSH1 4C |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
251587_at |
AT3G58080
|
unknown |
0.231 |
0.030 |
| 2 |
245183_at |
AT5G12440
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.192 |
-0.275 |
| 3 |
259833_at |
AT1G69560
|
ATMYB105, MYB DOMAIN PROTEIN 105, LOF2, LATERAL ORGAN FUSION 2, MYB105, myb domain protein 105 |
0.186 |
-0.017 |
| 4 |
250635_at |
AT5G07510
|
ATGRP-4, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 4, ATGRP14, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 14, GRP-4, GLYCINE-RICH PROTEIN 4, GRP14, glycine-rich protein 14 |
0.180 |
0.028 |
| 5 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.175 |
-0.111 |
| 6 |
253652_at |
AT4G30040
|
[Eukaryotic aspartyl protease family protein] |
0.149 |
0.038 |
| 7 |
250327_at |
AT5G12050
|
unknown |
0.143 |
-0.252 |
| 8 |
254627_at |
AT4G18395
|
unknown |
0.142 |
-0.055 |
| 9 |
246022_at |
AT5G21110
|
unknown |
0.142 |
0.005 |
| 10 |
257934_at |
AT3G25420
|
scpl21, serine carboxypeptidase-like 21 |
0.139 |
0.032 |
| 11 |
251751_at |
AT3G55720
|
unknown |
0.135 |
0.034 |
| 12 |
248216_at |
AT5G53690
|
unknown |
0.132 |
0.036 |
| 13 |
262598_at |
AT1G15260
|
unknown |
0.119 |
-0.442 |
| 14 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.117 |
0.545 |
| 15 |
245224_at |
AT3G29796
|
unknown |
0.116 |
-0.005 |
| 16 |
267386_at |
AT2G44430
|
[DNA-binding bromodomain-containing protein] |
0.116 |
0.030 |
| 17 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.259 |
0.915 |
| 18 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.224 |
-0.051 |
| 19 |
264646_at |
AT1G08860
|
BON3, BONZAI 3 |
-0.188 |
0.022 |
| 20 |
251323_at |
AT3G61580
|
AtSLD1, SLD1, sphingoid LCB desaturase 1 |
-0.167 |
0.062 |
| 21 |
266868_at |
AT2G44630
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.148 |
-0.065 |
| 22 |
253504_at |
AT4G31960
|
unknown |
-0.147 |
0.103 |
| 23 |
250491_at |
AT5G09780
|
[Transcriptional factor B3 family protein] |
-0.147 |
-0.011 |
| 24 |
267164_at |
AT2G37700
|
[Fatty acid hydroxylase superfamily] |
-0.144 |
0.051 |
| 25 |
259101_at |
AT3G11640
|
unknown |
-0.144 |
0.043 |
| 26 |
247526_at |
AT5G61470
|
[C2H2-like zinc finger protein] |
-0.133 |
0.001 |
| 27 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.133 |
-0.073 |
| 28 |
248714_at |
AT5G48140
|
[Pectin lyase-like superfamily protein] |
-0.132 |
0.004 |
| 29 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
-0.125 |
0.019 |
| 30 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.123 |
0.121 |
| 31 |
258389_at |
AT3G15390
|
SDE5, silencing defective 5 |
-0.120 |
-0.081 |