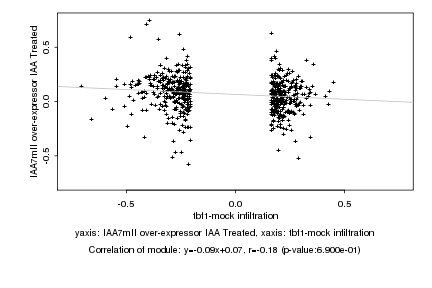
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tbf1-mock infiltration |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.448 |
0.180 |
| 2 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.428 |
0.100 |
| 3 |
245007_at |
ATCG00350
|
PSAA |
0.423 |
-0.024 |
| 4 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
0.410 |
0.054 |
| 5 |
264800_at |
AT1G08800
|
MyoB1, myosin binding protein 1 |
0.364 |
0.065 |
| 6 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
0.356 |
0.344 |
| 7 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.353 |
0.140 |
| 8 |
259816_at |
AT1G49850
|
[RING/U-box superfamily protein] |
0.344 |
-0.331 |
| 9 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.343 |
-0.028 |
| 10 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.343 |
0.092 |
| 11 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
0.338 |
0.076 |
| 12 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
0.335 |
0.114 |
| 13 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.333 |
0.051 |
| 14 |
266990_at |
AT2G39190
|
ATATH8 |
0.323 |
0.136 |
| 15 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
0.322 |
0.037 |
| 16 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.322 |
0.378 |
| 17 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
0.316 |
0.068 |
| 18 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.311 |
-0.115 |
| 19 |
261451_at |
AT1G21060
|
unknown |
0.308 |
-0.028 |
| 20 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.307 |
0.193 |
| 21 |
253322_at |
AT4G33980
|
unknown |
0.303 |
0.071 |
| 22 |
249614_at |
AT5G37300
|
WSD1 |
0.300 |
0.139 |
| 23 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.299 |
0.186 |
| 24 |
266340_at |
AT2G01480
|
[O-fucosyltransferase family protein] |
0.298 |
0.123 |
| 25 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.298 |
-0.003 |
| 26 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.294 |
0.141 |
| 27 |
250585_at |
AT5G07620
|
[Protein kinase superfamily protein] |
0.292 |
0.123 |
| 28 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.291 |
-0.082 |
| 29 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.290 |
-0.013 |
| 30 |
264238_at |
AT1G54740
|
unknown |
0.289 |
0.233 |
| 31 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.286 |
-0.516 |
| 32 |
260688_at |
AT1G17665
|
unknown |
0.285 |
-0.029 |
| 33 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.283 |
0.079 |
| 34 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
0.280 |
0.039 |
| 35 |
246325_at |
AT1G16540
|
ABA3, ABA DEFICIENT 3, ACI2, ALTERED CHLOROPLAST IMPORT 2, ATABA3, AtLOS5, LOS5, LOW OSMOTIC STRESS 5, SIR3, SIRTINOL RESISTANT 3 |
0.278 |
0.211 |
| 36 |
267102_at |
AT2G41500
|
EMB2776, LIS, LACHESIS |
0.278 |
-0.042 |
| 37 |
259113_at |
AT3G05510
|
[Phospholipid/glycerol acyltransferase family protein] |
0.277 |
-0.025 |
| 38 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.277 |
0.173 |
| 39 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.276 |
0.025 |
| 40 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.276 |
0.127 |
| 41 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.276 |
-0.026 |
| 42 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.274 |
0.068 |
| 43 |
251646_at |
AT3G57780
|
unknown |
0.274 |
-0.020 |
| 44 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.273 |
0.106 |
| 45 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.272 |
0.140 |
| 46 |
260081_at |
AT1G78170
|
unknown |
0.271 |
0.153 |
| 47 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.271 |
0.134 |
| 48 |
267019_at |
AT2G39130
|
[Transmembrane amino acid transporter family protein] |
0.271 |
-0.359 |
| 49 |
267511_at |
AT2G45670
|
[calcineurin B subunit-related] |
0.270 |
0.002 |
| 50 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.269 |
0.056 |
| 51 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.269 |
-0.103 |
| 52 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
0.269 |
-0.115 |
| 53 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
0.268 |
0.024 |
| 54 |
247676_at |
AT5G59980
|
AtRPP30, GAF1, GAMETOPHYTE DEFECTIVE 1, RPP30 |
0.266 |
0.061 |
| 55 |
251476_at |
AT3G59670
|
unknown |
0.265 |
0.201 |
| 56 |
254664_at |
AT4G18300
|
[Trimeric LpxA-like enzyme] |
0.264 |
0.008 |
| 57 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.263 |
-0.093 |
| 58 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.260 |
0.178 |
| 59 |
261205_at |
AT1G12790
|
unknown |
0.259 |
0.278 |
| 60 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
0.257 |
-0.100 |
| 61 |
251474_at |
AT3G59630
|
[diphthamide synthesis DPH2 family protein] |
0.257 |
0.169 |
| 62 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
0.256 |
0.137 |
| 63 |
264324_at |
AT1G04160
|
ATXIB, ARABIDOPSIS THALIANA MYOSIN XI B, XI-8, MYOSIN XI-8, XI-B, MYOSIN XI B, XIB, myosin XI B |
0.256 |
0.147 |
| 64 |
258509_at |
AT3G06620
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.255 |
0.004 |
| 65 |
253881_at |
AT4G27640
|
[ARM repeat superfamily protein] |
0.254 |
0.021 |
| 66 |
266793_at |
AT2G03060
|
AGL30, AGAMOUS-like 30 |
0.253 |
-0.086 |
| 67 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.253 |
-0.265 |
| 68 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.248 |
0.127 |
| 69 |
247522_at |
AT5G61340
|
unknown |
0.248 |
-0.017 |
| 70 |
262219_at |
AT1G74750
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.248 |
0.021 |
| 71 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.247 |
0.127 |
| 72 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.246 |
-0.213 |
| 73 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.245 |
-0.010 |
| 74 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
0.245 |
0.040 |
| 75 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.244 |
-0.102 |
| 76 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.244 |
0.260 |
| 77 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
0.243 |
0.299 |
| 78 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.242 |
-0.088 |
| 79 |
245587_at |
AT4G15020
|
[hAT transposon superfamily] |
0.241 |
0.090 |
| 80 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.241 |
0.192 |
| 81 |
261387_at |
AT1G05410
|
unknown |
0.241 |
-0.170 |
| 82 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.240 |
-0.081 |
| 83 |
262006_at |
AT1G64570
|
DUO3, DUO POLLEN 3 |
0.239 |
0.106 |
| 84 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.238 |
0.045 |
| 85 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.236 |
0.259 |
| 86 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.235 |
-0.096 |
| 87 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
0.235 |
-0.090 |
| 88 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.234 |
0.194 |
| 89 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.232 |
0.244 |
| 90 |
261460_at |
AT1G07880
|
ATMPK13 |
0.232 |
-0.250 |
| 91 |
249702_at |
AT5G35570
|
[O-fucosyltransferase family protein] |
0.232 |
0.012 |
| 92 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.229 |
-0.099 |
| 93 |
247751_at |
AT5G59050
|
unknown |
0.228 |
0.143 |
| 94 |
265875_at |
AT2G01690
|
[ARM repeat superfamily protein] |
0.227 |
-0.005 |
| 95 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.225 |
0.086 |
| 96 |
250828_at |
AT5G05250
|
unknown |
0.225 |
0.147 |
| 97 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.225 |
-0.011 |
| 98 |
256192_at |
AT1G30110
|
ATNUDX25, nudix hydrolase homolog 25, NUDX25, nudix hydrolase homolog 25 |
0.224 |
-0.020 |
| 99 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.224 |
-0.021 |
| 100 |
252659_at |
AT3G44430
|
unknown |
0.222 |
0.237 |
| 101 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.222 |
0.147 |
| 102 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.222 |
-0.023 |
| 103 |
249277_at |
AT5G41890
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.221 |
0.072 |
| 104 |
249763_at |
AT5G24010
|
[Protein kinase superfamily protein] |
0.221 |
-0.135 |
| 105 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
0.221 |
0.099 |
| 106 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.221 |
0.062 |
| 107 |
257815_at |
AT3G25130
|
unknown |
0.220 |
-0.246 |
| 108 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.220 |
-0.146 |
| 109 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.220 |
0.005 |
| 110 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.220 |
-0.301 |
| 111 |
265373_at |
AT2G06510
|
ATRPA1A, ARABIDOPSIS THALIANA REPLICATION PROTEIN A 1A, ATRPA70A, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT A, RPA1A, replication protein A 1A, RPA70A, RPA70-KDA SUBUNIT A |
0.219 |
0.024 |
| 112 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.218 |
0.099 |
| 113 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
0.218 |
0.199 |
| 114 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.218 |
0.144 |
| 115 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.217 |
-0.147 |
| 116 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.217 |
-0.053 |
| 117 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.217 |
0.101 |
| 118 |
250823_at |
AT5G05180
|
unknown |
0.216 |
-0.120 |
| 119 |
257825_at |
AT3G26700
|
[Protein kinase superfamily protein] |
0.216 |
-0.048 |
| 120 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.215 |
0.148 |
| 121 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.214 |
0.089 |
| 122 |
262071_at |
AT1G59510
|
CF9 |
0.214 |
-0.166 |
| 123 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
0.214 |
0.019 |
| 124 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.213 |
0.286 |
| 125 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.213 |
-0.006 |
| 126 |
264286_at |
AT1G61870
|
PPR336, pentatricopeptide repeat 336 |
0.212 |
0.041 |
| 127 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.212 |
0.182 |
| 128 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.212 |
-0.167 |
| 129 |
263163_at |
AT1G03160
|
FZL, FZO-like |
0.212 |
-0.069 |
| 130 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.211 |
0.037 |
| 131 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.210 |
-0.004 |
| 132 |
250710_at |
AT5G06100
|
ATMYB33, MYB33, myb domain protein 33 |
0.210 |
0.181 |
| 133 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.210 |
-0.130 |
| 134 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.208 |
0.183 |
| 135 |
251375_at |
AT3G60410
|
unknown |
0.207 |
0.024 |
| 136 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.206 |
-0.230 |
| 137 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.206 |
0.312 |
| 138 |
255673_at |
AT4G00340
|
RLK4, receptor-like protein kinase 4 |
0.206 |
0.122 |
| 139 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
0.205 |
-0.085 |
| 140 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.205 |
0.105 |
| 141 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.205 |
0.069 |
| 142 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.205 |
-0.209 |
| 143 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
0.205 |
0.006 |
| 144 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.204 |
0.083 |
| 145 |
265395_at |
AT2G20850
|
SRF1, STRUBBELIG-receptor family 1 |
0.204 |
-0.025 |
| 146 |
251224_at |
AT3G62620
|
[sucrose-phosphatase-related] |
0.204 |
0.057 |
| 147 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.204 |
0.051 |
| 148 |
264625_at |
AT1G09020
|
ATSNF4, SNF4, homolog of yeast sucrose nonfermenting 4 |
0.204 |
0.086 |
| 149 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.203 |
-0.113 |
| 150 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.203 |
0.168 |
| 151 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.203 |
0.088 |
| 152 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.203 |
0.217 |
| 153 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.202 |
-0.126 |
| 154 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.202 |
0.106 |
| 155 |
260142_at |
AT1G71900
|
unknown |
0.202 |
0.098 |
| 156 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
0.201 |
0.109 |
| 157 |
258899_at |
AT3G05675
|
[BTB/POZ domain-containing protein] |
0.201 |
-0.148 |
| 158 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.201 |
0.170 |
| 159 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
0.201 |
0.014 |
| 160 |
246762_at |
AT5G27620
|
CYCH;1, cyclin H;1 |
0.201 |
-0.051 |
| 161 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
0.199 |
0.287 |
| 162 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.199 |
0.105 |
| 163 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.198 |
0.350 |
| 164 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.198 |
0.287 |
| 165 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
0.198 |
0.229 |
| 166 |
260655_at |
AT1G19320
|
[Pathogenesis-related thaumatin superfamily protein] |
0.197 |
0.104 |
| 167 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.196 |
0.111 |
| 168 |
266124_at |
AT2G45080
|
cycp3;1, cyclin p3;1 |
0.196 |
0.234 |
| 169 |
262531_at |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
0.195 |
0.302 |
| 170 |
267475_at |
AT2G02730
|
unknown |
0.195 |
-0.000 |
| 171 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.195 |
0.141 |
| 172 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.195 |
-0.142 |
| 173 |
259322_at |
AT3G05270
|
unknown |
0.195 |
-0.022 |
| 174 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.195 |
0.151 |
| 175 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.195 |
-0.175 |
| 176 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.195 |
0.092 |
| 177 |
265098_at |
AT1G04010
|
ATPSAT1, ERP1, enhanced response to Phytophthora 1, PSAT1, phospholipid sterol acyl transferase 1 |
0.194 |
0.006 |
| 178 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.194 |
-0.090 |
| 179 |
245909_at |
AT5G09380
|
[RNA polymerase III RPC4] |
0.194 |
-0.445 |
| 180 |
257829_at |
AT3G26680
|
ATSNM1, SENSITIVE TO NITROGEN MUSTARD 1, SNM1, SENSITIVE TO NITROGEN MUSTARD 1 |
0.194 |
-0.020 |
| 181 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.193 |
-0.004 |
| 182 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.192 |
0.244 |
| 183 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.192 |
-0.003 |
| 184 |
247605_at |
AT5G60970
|
TCP5, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 |
0.192 |
0.011 |
| 185 |
246583_at |
AT5G14720
|
[Protein kinase superfamily protein] |
0.192 |
0.098 |
| 186 |
261467_at |
AT1G28520
|
ATVOZ1, VASCULAR PLANT ONE ZINC FINGER PROTEIN, VOZ1, vascular plant one zinc finger protein |
0.192 |
-0.048 |
| 187 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.192 |
0.010 |
| 188 |
247378_at |
AT5G63120
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.191 |
0.164 |
| 189 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.191 |
-0.043 |
| 190 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.191 |
-0.109 |
| 191 |
255497_at |
AT4G02590
|
UNE12, unfertilized embryo sac 12 |
0.191 |
0.002 |
| 192 |
259973_at |
AT1G76630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.191 |
-0.223 |
| 193 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
0.190 |
0.237 |
| 194 |
247276_at |
AT5G64390
|
HEN4, HUA ENHANCER 4 |
0.190 |
0.112 |
| 195 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.190 |
0.152 |
| 196 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.190 |
0.171 |
| 197 |
251796_at |
AT3G55360
|
ATTSC13, CER10, ECERIFERUM 10, ECR, ENOYL-COA REDUCTASE, TSC13 |
0.190 |
0.171 |
| 198 |
259373_at |
AT1G69160
|
unknown |
0.190 |
0.117 |
| 199 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.189 |
0.112 |
| 200 |
263314_at |
AT2G05760
|
[Xanthine/uracil permease family protein] |
0.189 |
0.016 |
| 201 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
0.189 |
0.035 |
| 202 |
250571_at |
AT5G08200
|
[peptidoglycan-binding LysM domain-containing protein] |
0.188 |
-0.023 |
| 203 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.188 |
0.069 |
| 204 |
264424_at |
AT1G61740
|
[Sulfite exporter TauE/SafE family protein] |
0.188 |
-0.024 |
| 205 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.188 |
0.024 |
| 206 |
250826_at |
AT5G05220
|
unknown |
0.188 |
0.003 |
| 207 |
258877_at |
AT3G03220
|
ATEXP13, EXPANSIN 13, ATEXPA13, expansin A13, ATHEXP ALPHA 1.22, EXP13, EXPA13, expansin A13 |
0.188 |
-0.026 |
| 208 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.188 |
0.004 |
| 209 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.188 |
0.468 |
| 210 |
262849_at |
AT1G14710
|
[hydroxyproline-rich glycoprotein family protein] |
0.187 |
-0.056 |
| 211 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.187 |
0.065 |
| 212 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.186 |
0.142 |
| 213 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
0.185 |
-0.048 |
| 214 |
261127_at |
AT1G04790
|
[RING/U-box superfamily protein] |
0.185 |
0.011 |
| 215 |
257709_at |
AT3G27325
|
[hydrolases, acting on ester bonds] |
0.185 |
0.053 |
| 216 |
253402_at |
AT4G32880
|
ATHB-8, homeobox gene 8, ATHB8, HB-8, homeobox gene 8 |
0.185 |
0.026 |
| 217 |
245112_at |
AT2G41540
|
GPDHC1 |
0.185 |
0.167 |
| 218 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.184 |
-0.081 |
| 219 |
258978_at |
AT3G09320
|
[DHHC-type zinc finger family protein] |
0.184 |
0.037 |
| 220 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.184 |
-0.089 |
| 221 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.184 |
0.151 |
| 222 |
250449_at |
AT5G10830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.183 |
0.083 |
| 223 |
264961_at |
AT1G76950
|
PRAF1 |
0.183 |
-0.028 |
| 224 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.183 |
0.088 |
| 225 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.183 |
0.092 |
| 226 |
250939_at |
AT5G03040
|
iqd2, IQ-domain 2 |
0.182 |
-0.069 |
| 227 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.182 |
0.239 |
| 228 |
250066_at |
AT5G17930
|
[MIF4G domain-containing protein / MA3 domain-containing protein] |
0.181 |
0.135 |
| 229 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.181 |
0.405 |
| 230 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.181 |
0.094 |
| 231 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
0.181 |
0.221 |
| 232 |
251914_at |
AT3G53930
|
[Protein kinase superfamily protein] |
0.180 |
0.264 |
| 233 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
0.180 |
-0.043 |
| 234 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.179 |
-0.015 |
| 235 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
0.178 |
0.048 |
| 236 |
258260_at |
AT3G26850
|
[histone-lysine N-methyltransferases] |
0.178 |
-0.087 |
| 237 |
245353_at |
AT4G16000
|
unknown |
0.178 |
0.139 |
| 238 |
245290_at |
AT4G16490
|
[ARM repeat superfamily protein] |
0.178 |
-0.100 |
| 239 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.178 |
0.156 |
| 240 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
0.177 |
-0.039 |
| 241 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
0.177 |
0.091 |
| 242 |
249831_at |
AT5G23340
|
[RNI-like superfamily protein] |
0.177 |
0.092 |
| 243 |
248053_at |
AT5G55810
|
AtNMNAT, nicotinate/nicotinamide mononucleotide adenyltransferase, NMNAT, nicotinate/nicotinamide mononucleotide adenyltransferase |
0.176 |
-0.120 |
| 244 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.176 |
0.421 |
| 245 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.176 |
-0.090 |
| 246 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
0.175 |
0.243 |
| 247 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.175 |
-0.076 |
| 248 |
246964_at |
AT5G24830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.175 |
0.063 |
| 249 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.174 |
0.158 |
| 250 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.174 |
0.071 |
| 251 |
252471_at |
AT3G46820
|
TOPP5, type one serine/threonine protein phosphatase 5 |
0.174 |
0.004 |
| 252 |
247633_at |
AT5G60470
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.174 |
0.169 |
| 253 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.173 |
0.193 |
| 254 |
260282_at |
AT1G80410
|
EMB2753, EMBRYO DEFECTIVE 2753, OMA, OMISHA |
0.173 |
0.013 |
| 255 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.173 |
-0.083 |
| 256 |
257260_at |
AT3G22104
|
[Phototropic-responsive NPH3 family protein] |
0.173 |
-0.014 |
| 257 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.173 |
0.179 |
| 258 |
245138_at |
AT2G45190
|
AFO, ABNORMAL FLORAL ORGANS, FIL, FILAMENTOUS FLOWER, YAB1, YABBY1 |
0.173 |
0.104 |
| 259 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.172 |
0.080 |
| 260 |
247012_at |
AT5G67580
|
ATTBP3, ATTRB2, TELOMERE REPEAT BINDING FACTOR 2, TBP3, TELOMERE-BINDING PROTEIN 3, TRB2 |
0.172 |
-0.116 |
| 261 |
246593_at |
AT5G14790
|
[ARM repeat superfamily protein] |
0.172 |
0.083 |
| 262 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
0.172 |
-0.018 |
| 263 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.172 |
0.030 |
| 264 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.172 |
-0.182 |
| 265 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
0.172 |
-0.179 |
| 266 |
246141_at |
AT5G19920
|
[Transducin/WD40 repeat-like superfamily protein] |
0.171 |
0.100 |
| 267 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.171 |
0.159 |
| 268 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.171 |
0.068 |
| 269 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.171 |
-0.247 |
| 270 |
249991_at |
AT5G18550
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.171 |
-0.171 |
| 271 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.170 |
0.188 |
| 272 |
251882_at |
AT3G54140
|
AtNPF8.1, ATPTR1, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 1, NPF8.1, NRT1/ PTR family 8.1, PTR1, peptide transporter 1 |
0.169 |
0.045 |
| 273 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
0.169 |
0.228 |
| 274 |
255821_at |
AT2G40570
|
[initiator tRNA phosphoribosyl transferase family protein] |
0.169 |
-0.081 |
| 275 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
0.168 |
0.245 |
| 276 |
246399_at |
AT1G58110
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
0.168 |
-0.067 |
| 277 |
248023_at |
AT5G56450
|
PM-ANT |
0.168 |
0.075 |
| 278 |
267616_at |
AT2G26680
|
unknown |
0.168 |
-0.123 |
| 279 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.168 |
0.245 |
| 280 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
0.166 |
0.112 |
| 281 |
246497_at |
AT5G16220
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.166 |
-0.072 |
| 282 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.165 |
0.012 |
| 283 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.165 |
-0.113 |
| 284 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
0.164 |
0.384 |
| 285 |
266626_at |
AT2G35360
|
[ubiquitin family protein] |
0.164 |
-0.215 |
| 286 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.164 |
0.182 |
| 287 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
0.164 |
0.402 |
| 288 |
247631_at |
AT5G60440
|
AGL62, AGAMOUS-like 62 |
0.164 |
-0.258 |
| 289 |
248873_at |
AT5G46450
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.164 |
0.099 |
| 290 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.163 |
0.170 |
| 291 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.163 |
-0.005 |
| 292 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.163 |
0.077 |
| 293 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.163 |
-0.026 |
| 294 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.163 |
-0.070 |
| 295 |
251849_at |
AT3G54610
|
BGT, BIG TOP, GCN5, general control non-repressible 5, HAC3, HAG01, HAG1, histone acetyltransferase of the GNAT family 1, HAT1, HISTONE ACETYLTRANSFERASE 1 |
0.163 |
-0.210 |
| 296 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.162 |
0.105 |
| 297 |
265078_at |
AT1G55500
|
ECT4, evolutionarily conserved C-terminal region 4 |
0.162 |
-0.092 |
| 298 |
266349_at |
AT2G01460
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.162 |
0.248 |
| 299 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.162 |
0.633 |
| 300 |
245431_at |
AT4G17080
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.162 |
0.071 |
| 301 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.706 |
0.139 |
| 302 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-0.660 |
-0.160 |
| 303 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.598 |
0.037 |
| 304 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.568 |
-0.067 |
| 305 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.548 |
0.141 |
| 306 |
253832_at |
AT4G27654
|
unknown |
-0.546 |
0.208 |
| 307 |
245755_at |
AT1G35210
|
unknown |
-0.511 |
0.159 |
| 308 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.509 |
-0.043 |
| 309 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.499 |
-0.221 |
| 310 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.487 |
0.051 |
| 311 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.484 |
0.594 |
| 312 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
-0.483 |
0.163 |
| 313 |
252073_at |
AT3G51750
|
unknown |
-0.480 |
-0.117 |
| 314 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.478 |
0.138 |
| 315 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.474 |
0.186 |
| 316 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.468 |
0.014 |
| 317 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.459 |
0.152 |
| 318 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.453 |
0.164 |
| 319 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.448 |
0.172 |
| 320 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.447 |
0.201 |
| 321 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.446 |
0.165 |
| 322 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.445 |
0.074 |
| 323 |
259850_at |
AT1G72240
|
unknown |
-0.444 |
0.185 |
| 324 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.442 |
0.170 |
| 325 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
-0.431 |
0.088 |
| 326 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.429 |
0.037 |
| 327 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
-0.427 |
0.091 |
| 328 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.427 |
-0.081 |
| 329 |
264958_at |
AT1G76960
|
unknown |
-0.421 |
0.039 |
| 330 |
251610_at |
AT3G57930
|
unknown |
-0.421 |
-0.330 |
| 331 |
256442_at |
AT3G10930
|
unknown |
-0.419 |
0.093 |
| 332 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.413 |
0.224 |
| 333 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-0.412 |
0.228 |
| 334 |
252346_at |
AT3G48650
|
[pseudogene] |
-0.411 |
0.074 |
| 335 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-0.410 |
0.718 |
| 336 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
-0.408 |
-0.082 |
| 337 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-0.401 |
0.234 |
| 338 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.397 |
0.208 |
| 339 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.395 |
0.227 |
| 340 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
-0.395 |
0.755 |
| 341 |
246200_at |
AT4G37240
|
unknown |
-0.394 |
0.247 |
| 342 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.393 |
0.175 |
| 343 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.390 |
0.154 |
| 344 |
249522_at |
AT5G38700
|
unknown |
-0.388 |
-0.026 |
| 345 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.386 |
0.154 |
| 346 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.380 |
0.087 |
| 347 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.378 |
0.204 |
| 348 |
253796_at |
AT4G28460
|
unknown |
-0.378 |
0.140 |
| 349 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.375 |
0.206 |
| 350 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.373 |
0.065 |
| 351 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.372 |
0.244 |
| 352 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
-0.369 |
-0.031 |
| 353 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.365 |
0.230 |
| 354 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.361 |
0.128 |
| 355 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.360 |
0.044 |
| 356 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.358 |
0.144 |
| 357 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.358 |
0.276 |
| 358 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
-0.357 |
0.047 |
| 359 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.354 |
0.578 |
| 360 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.354 |
0.191 |
| 361 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.351 |
-0.067 |
| 362 |
263182_at |
AT1G05575
|
unknown |
-0.348 |
0.333 |
| 363 |
250796_at |
AT5G05300
|
unknown |
-0.348 |
0.078 |
| 364 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.347 |
0.241 |
| 365 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
-0.347 |
0.333 |
| 366 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.345 |
0.196 |
| 367 |
267344_at |
AT2G44230
|
unknown |
-0.344 |
-0.024 |
| 368 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.344 |
0.176 |
| 369 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.337 |
0.154 |
| 370 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.337 |
0.259 |
| 371 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.337 |
0.214 |
| 372 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
-0.337 |
0.122 |
| 373 |
253859_at |
AT4G27657
|
unknown |
-0.336 |
-0.041 |
| 374 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.333 |
0.198 |
| 375 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.331 |
0.149 |
| 376 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.331 |
0.225 |
| 377 |
256569_at |
AT3G19550
|
unknown |
-0.330 |
0.099 |
| 378 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.329 |
0.052 |
| 379 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.329 |
0.308 |
| 380 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.328 |
0.129 |
| 381 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.327 |
0.004 |
| 382 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
-0.326 |
-0.081 |
| 383 |
250098_at |
AT5G17350
|
unknown |
-0.324 |
0.140 |
| 384 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.324 |
-0.028 |
| 385 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.324 |
-0.009 |
| 386 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.323 |
0.138 |
| 387 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.322 |
0.218 |
| 388 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.319 |
0.034 |
| 389 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.319 |
-0.044 |
| 390 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.319 |
0.239 |
| 391 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.318 |
-0.114 |
| 392 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.317 |
0.079 |
| 393 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.317 |
0.404 |
| 394 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.315 |
0.160 |
| 395 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.315 |
0.174 |
| 396 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.315 |
0.049 |
| 397 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.314 |
0.157 |
| 398 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.314 |
-0.198 |
| 399 |
248509_at |
AT5G50335
|
unknown |
-0.314 |
0.110 |
| 400 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.313 |
0.127 |
| 401 |
259385_at |
AT1G13470
|
unknown |
-0.311 |
0.206 |
| 402 |
266396_at |
AT2G38790
|
unknown |
-0.309 |
0.296 |
| 403 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.308 |
0.187 |
| 404 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.307 |
-0.153 |
| 405 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
-0.307 |
0.104 |
| 406 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-0.306 |
0.185 |
| 407 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
-0.305 |
0.182 |
| 408 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.304 |
0.283 |
| 409 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.304 |
0.129 |
| 410 |
260005_at |
AT1G67920
|
unknown |
-0.304 |
0.052 |
| 411 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
-0.301 |
0.038 |
| 412 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.300 |
0.209 |
| 413 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.300 |
0.108 |
| 414 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.298 |
0.037 |
| 415 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.297 |
0.127 |
| 416 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.295 |
0.102 |
| 417 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.294 |
0.277 |
| 418 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.292 |
0.231 |
| 419 |
246842_at |
AT5G26731
|
unknown |
-0.292 |
-0.201 |
| 420 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
-0.291 |
-0.147 |
| 421 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.291 |
-0.515 |
| 422 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.291 |
0.177 |
| 423 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.291 |
0.112 |
| 424 |
252023_at |
AT3G52920
|
unknown |
-0.291 |
0.167 |
| 425 |
267409_at |
AT2G34910
|
unknown |
-0.290 |
-0.195 |
| 426 |
249932_at |
AT5G22390
|
unknown |
-0.288 |
0.038 |
| 427 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.288 |
0.086 |
| 428 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.288 |
-0.365 |
| 429 |
265189_at |
AT1G23840
|
unknown |
-0.287 |
0.095 |
| 430 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.287 |
0.052 |
| 431 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
-0.287 |
0.082 |
| 432 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.286 |
0.180 |
| 433 |
253044_at |
AT4G37290
|
unknown |
-0.284 |
0.076 |
| 434 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.284 |
-0.072 |
| 435 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.284 |
0.006 |
| 436 |
258351_at |
AT3G17700
|
ATCNGC20, CYCLIC NUCLEOTIDE-GATED CHANNEL 20, CNBT1, cyclic nucleotide-binding transporter 1, CNGC20, cyclic nucleotide gated channel 20 |
-0.283 |
0.228 |
| 437 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
-0.283 |
-0.002 |
| 438 |
248775_at |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
-0.282 |
0.143 |
| 439 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.282 |
0.238 |
| 440 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
-0.281 |
0.179 |
| 441 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
-0.280 |
-0.210 |
| 442 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.278 |
-0.467 |
| 443 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.278 |
0.237 |
| 444 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.276 |
0.065 |
| 445 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
-0.276 |
0.069 |
| 446 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.275 |
-0.009 |
| 447 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.275 |
-0.068 |
| 448 |
261921_at |
AT1G65900
|
unknown |
-0.274 |
0.106 |
| 449 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.274 |
0.018 |
| 450 |
253827_at |
AT4G28085
|
unknown |
-0.273 |
0.282 |
| 451 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
-0.273 |
0.069 |
| 452 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.273 |
-0.049 |
| 453 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.273 |
-0.066 |
| 454 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.272 |
0.337 |
| 455 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.270 |
0.200 |
| 456 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.269 |
0.127 |
| 457 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.269 |
0.127 |
| 458 |
265993_at |
AT2G24160
|
[pseudogene] |
-0.269 |
0.191 |
| 459 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.268 |
0.273 |
| 460 |
246370_at |
AT1G51920
|
unknown |
-0.267 |
0.000 |
| 461 |
250936_at |
AT5G03120
|
unknown |
-0.266 |
0.091 |
| 462 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.266 |
-0.149 |
| 463 |
262211_at |
AT1G74930
|
ORA47 |
-0.265 |
0.138 |
| 464 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.265 |
0.177 |
| 465 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.265 |
0.030 |
| 466 |
250575_at |
AT5G08240
|
unknown |
-0.265 |
0.089 |
| 467 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.264 |
0.102 |
| 468 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.263 |
0.125 |
| 469 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.263 |
0.133 |
| 470 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.262 |
0.286 |
| 471 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.262 |
0.346 |
| 472 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.261 |
0.067 |
| 473 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.261 |
0.028 |
| 474 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.261 |
0.122 |
| 475 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.261 |
0.198 |
| 476 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.260 |
0.288 |
| 477 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.260 |
0.158 |
| 478 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.257 |
-0.263 |
| 479 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.257 |
0.624 |
| 480 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.257 |
-0.013 |
| 481 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.255 |
-0.134 |
| 482 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.254 |
0.175 |
| 483 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.253 |
0.227 |
| 484 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.253 |
0.113 |
| 485 |
257956_at |
AT3G25400
|
unknown |
-0.253 |
-0.091 |
| 486 |
262072_at |
AT1G59590
|
ZCF37 |
-0.252 |
-0.110 |
| 487 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.252 |
0.081 |
| 488 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.251 |
0.186 |
| 489 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.250 |
0.080 |
| 490 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
-0.249 |
0.187 |
| 491 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.248 |
-0.022 |
| 492 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.248 |
-0.465 |
| 493 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
-0.247 |
-0.033 |
| 494 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.247 |
0.131 |
| 495 |
261381_at |
AT1G05460
|
SDE3, SILENCING DEFECTIVE |
-0.247 |
0.069 |
| 496 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
-0.246 |
0.109 |
| 497 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.245 |
0.040 |
| 498 |
261221_at |
AT1G19960
|
unknown |
-0.245 |
0.140 |
| 499 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.245 |
0.138 |
| 500 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.245 |
-0.054 |
| 501 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.244 |
0.135 |
| 502 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
-0.243 |
0.336 |
| 503 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.243 |
-0.220 |
| 504 |
263083_at |
AT2G27190
|
ATPAP1, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 1, ATPAP12, PURPLE ACID PHOSPHATASE 12, PAP1, PURPLE ACID PHOSPHATASE 1, PAP12, purple acid phosphatase 12 |
-0.243 |
0.029 |
| 505 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.243 |
0.084 |
| 506 |
258203_at |
AT3G13950
|
unknown |
-0.242 |
0.486 |
| 507 |
257925_at |
AT3G23170
|
unknown |
-0.242 |
0.273 |
| 508 |
264201_at |
AT1G22630
|
unknown |
-0.242 |
-0.042 |
| 509 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.242 |
-0.095 |
| 510 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-0.242 |
0.040 |
| 511 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
-0.242 |
0.019 |
| 512 |
251142_at |
AT5G01015
|
unknown |
-0.242 |
0.102 |
| 513 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
-0.241 |
0.169 |
| 514 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.240 |
0.226 |
| 515 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
-0.240 |
0.135 |
| 516 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.239 |
0.054 |
| 517 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.239 |
-0.131 |
| 518 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.239 |
0.071 |
| 519 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.239 |
-0.281 |
| 520 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.238 |
0.115 |
| 521 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
-0.237 |
-0.107 |
| 522 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.236 |
-0.004 |
| 523 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.235 |
0.094 |
| 524 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.235 |
0.029 |
| 525 |
257875_at |
AT3G17120
|
unknown |
-0.235 |
0.153 |
| 526 |
265737_at |
AT2G01180
|
ATLPP1, ATPAP1, phosphatidic acid phosphatase 1, LPP1, LIPID PHOSPHATE PHOSPHATASE 1, PAP1, phosphatidic acid phosphatase 1 |
-0.235 |
0.161 |
| 527 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.235 |
0.050 |
| 528 |
265184_at |
AT1G23710
|
unknown |
-0.234 |
0.064 |
| 529 |
263972_at |
AT2G42760
|
unknown |
-0.234 |
0.071 |
| 530 |
245209_at |
AT5G12340
|
unknown |
-0.233 |
0.175 |
| 531 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.233 |
-0.161 |
| 532 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.232 |
-0.168 |
| 533 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.232 |
0.112 |
| 534 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.231 |
0.016 |
| 535 |
262085_at |
AT1G56060
|
unknown |
-0.231 |
0.321 |
| 536 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.230 |
0.084 |
| 537 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.230 |
0.275 |
| 538 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.229 |
0.340 |
| 539 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
-0.229 |
-0.234 |
| 540 |
252345_at |
AT3G48640
|
unknown |
-0.229 |
0.204 |
| 541 |
252215_at |
AT3G50240
|
KICP-02 |
-0.229 |
-0.065 |
| 542 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.228 |
0.086 |
| 543 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.228 |
0.182 |
| 544 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
-0.226 |
0.382 |
| 545 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
-0.226 |
0.213 |
| 546 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
-0.226 |
0.199 |
| 547 |
262286_at |
AT1G68585
|
unknown |
-0.226 |
-0.078 |
| 548 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.225 |
0.146 |
| 549 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.224 |
-0.077 |
| 550 |
260735_at |
AT1G17610
|
CHS1, CHILLING SENSITIVE 1 |
-0.224 |
0.050 |
| 551 |
259661_at |
AT1G55265
|
unknown |
-0.224 |
0.160 |
| 552 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
-0.224 |
-0.005 |
| 553 |
255884_at |
AT1G20310
|
unknown |
-0.223 |
0.033 |
| 554 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.222 |
0.148 |
| 555 |
259489_at |
AT1G15790
|
unknown |
-0.222 |
-0.049 |
| 556 |
251332_at |
AT3G61670
|
unknown |
-0.222 |
-0.027 |
| 557 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.222 |
0.239 |
| 558 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.222 |
0.160 |
| 559 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.221 |
0.419 |
| 560 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.221 |
-0.052 |
| 561 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.221 |
0.156 |
| 562 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.221 |
0.292 |
| 563 |
249903_at |
AT5G22690
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.220 |
-0.113 |
| 564 |
249454_at |
AT5G39520
|
unknown |
-0.220 |
0.345 |
| 565 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
-0.220 |
-0.236 |
| 566 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.220 |
0.027 |
| 567 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.219 |
-0.063 |
| 568 |
261700_at |
AT1G32690
|
unknown |
-0.219 |
-0.020 |
| 569 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.219 |
0.077 |
| 570 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.219 |
0.090 |
| 571 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.218 |
0.162 |
| 572 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.218 |
0.019 |
| 573 |
261203_at |
AT1G12845
|
unknown |
-0.217 |
-0.086 |
| 574 |
264023_at |
AT2G21195
|
unknown |
-0.217 |
-0.231 |
| 575 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.217 |
0.144 |
| 576 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.217 |
0.312 |
| 577 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.217 |
-0.577 |
| 578 |
255912_at |
AT1G66960
|
LUP5 |
-0.215 |
0.100 |
| 579 |
249889_at |
AT5G22540
|
unknown |
-0.215 |
0.069 |
| 580 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.215 |
0.104 |
| 581 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.215 |
0.107 |
| 582 |
247740_at |
AT5G58940
|
CRCK1, calmodulin-binding receptor-like cytoplasmic kinase 1 |
-0.214 |
0.053 |
| 583 |
257545_at |
AT3G23200
|
[Uncharacterised protein family (UPF0497)] |
-0.214 |
-0.052 |
| 584 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.214 |
0.065 |
| 585 |
245776_at |
AT1G30260
|
unknown |
-0.213 |
0.103 |
| 586 |
255779_at |
AT1G18650
|
PDCB3, plasmodesmata callose-binding protein 3 |
-0.212 |
0.116 |
| 587 |
252548_at |
AT3G45850
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.212 |
0.036 |
| 588 |
247933_at |
AT5G56980
|
unknown |
-0.211 |
0.118 |
| 589 |
248715_at |
AT5G48290
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.211 |
-0.079 |
| 590 |
265276_at |
AT2G28400
|
unknown |
-0.211 |
0.266 |
| 591 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.210 |
-0.042 |
| 592 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.210 |
0.321 |
| 593 |
258134_at |
AT3G24530
|
[AAA-type ATPase family protein / ankyrin repeat family protein] |
-0.209 |
0.230 |
| 594 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
-0.209 |
0.111 |
| 595 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.208 |
0.158 |
| 596 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.208 |
0.212 |
| 597 |
251840_at |
AT3G54960
|
ATPDI1, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 1, ATPDIL1-3, PDI-like 1-3, PDI1, PROTEIN DISULFIDE ISOMERASE 1, PDIL1-3, PDI-like 1-3 |
-0.208 |
-0.024 |
| 598 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.208 |
-0.230 |
| 599 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
-0.208 |
0.005 |
| 600 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.207 |
-0.355 |