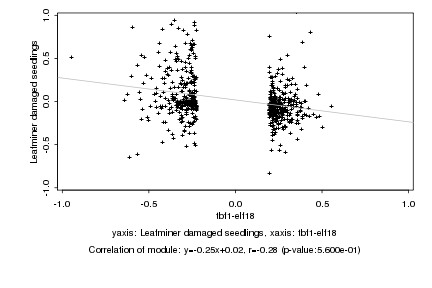
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tbf1-elf18 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
255688_at |
AT4G00660
|
ATRH8, RH8, RNAhelicase-like 8 |
0.553 |
-0.048 |
| 2 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.498 |
-0.297 |
| 3 |
255821_at |
AT2G40570
|
[initiator tRNA phosphoribosyl transferase family protein] |
0.481 |
-0.163 |
| 4 |
246219_at |
AT4G36760
|
APP1, aminopeptidase P1, ATAPP1, ARABIDOPSIS THALIANA AMINOPEPTIDASE P1 |
0.476 |
0.083 |
| 5 |
262693_at |
AT1G62780
|
unknown |
0.465 |
-0.174 |
| 6 |
266896_at |
AT2G45810
|
[DEA(D/H)-box RNA helicase family protein] |
0.450 |
-0.150 |
| 7 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.428 |
0.811 |
| 8 |
250865_at |
AT5G03900
|
[Iron-sulphur cluster biosynthesis family protein] |
0.427 |
-0.163 |
| 9 |
262415_at |
AT1G49400
|
emb1129, embryo defective 1129 |
0.418 |
-0.073 |
| 10 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.410 |
0.188 |
| 11 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
0.410 |
-0.151 |
| 12 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
0.401 |
0.009 |
| 13 |
260055_at |
AT1G78150
|
unknown |
0.398 |
-0.147 |
| 14 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.398 |
0.405 |
| 15 |
262531_at |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
0.394 |
0.006 |
| 16 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.384 |
-0.173 |
| 17 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.383 |
0.689 |
| 18 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.382 |
-0.186 |
| 19 |
262794_at |
AT1G13120
|
AtGLE1, A. thaliana homolog of yeast GLE1, GLE1 |
0.381 |
-0.064 |
| 20 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
0.377 |
-0.324 |
| 21 |
251465_at |
AT3G59360
|
ATUTR6, UDP-GALACTOSE TRANSPORTER 6, UTR6, UDP-galactose transporter 6 |
0.376 |
-0.012 |
| 22 |
251362_at |
AT3G61240
|
[DEA(D/H)-box RNA helicase family protein] |
0.375 |
-0.060 |
| 23 |
249592_at |
AT5G37890
|
[Protein with RING/U-box and TRAF-like domains] |
0.375 |
-0.099 |
| 24 |
264060_at |
AT2G27980
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.375 |
-0.107 |
| 25 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
0.366 |
0.000 |
| 26 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
0.359 |
-0.245 |
| 27 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.358 |
-0.438 |
| 28 |
246469_at |
AT5G17070
|
unknown |
0.356 |
0.141 |
| 29 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
0.356 |
-0.218 |
| 30 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.356 |
0.042 |
| 31 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.355 |
-0.159 |
| 32 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.355 |
-0.142 |
| 33 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
0.354 |
-0.123 |
| 34 |
255959_at |
AT1G21980
|
ATPIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1, ATPIPK1, PIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1 |
0.351 |
-0.002 |
| 35 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.351 |
1.042 |
| 36 |
249404_at |
AT5G40160
|
EMB139, EMBRYO DEFECTIVE 139, EMB506, embryo defective 506 |
0.351 |
-0.055 |
| 37 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
0.349 |
-0.026 |
| 38 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
0.348 |
-0.251 |
| 39 |
259071_at |
AT3G11650
|
NHL2, NDR1/HIN1-like 2 |
0.346 |
0.090 |
| 40 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.342 |
0.209 |
| 41 |
249926_at |
AT5G19180
|
ECR1, E1 C-terminal related 1 |
0.341 |
0.013 |
| 42 |
266130_at |
AT2G44980
|
ASG3, ALTERED SEED GERMINATION 3, CHR10, chromatin remodeling 10 |
0.336 |
0.059 |
| 43 |
251460_at |
AT3G59770
|
AtSAC9, ARABIDOPSIS THALIANA SUPPRESSOR OF ACTIN 9, SAC9, SUPPRESSOR OF ACTIN 9 |
0.335 |
0.003 |
| 44 |
255279_at |
AT4G04950
|
AtGRXS17, Arabidopsis thaliana monothiol glutaredoxin 17, GRXS17, monothiol glutaredoxin 17 |
0.333 |
-0.032 |
| 45 |
259564_at |
AT1G20540
|
[Transducin/WD40 repeat-like superfamily protein] |
0.331 |
-0.066 |
| 46 |
250739_at |
AT5G05740
|
ATEGY2, EGY2, ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
0.327 |
-0.202 |
| 47 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
0.325 |
0.277 |
| 48 |
249054_at |
AT5G44450
|
[methyltransferases] |
0.322 |
-0.044 |
| 49 |
267578_at |
AT2G30695
|
unknown |
0.321 |
-0.085 |
| 50 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.320 |
-0.197 |
| 51 |
257672_at |
AT3G20300
|
unknown |
0.320 |
0.031 |
| 52 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.319 |
0.207 |
| 53 |
261208_at |
AT1G12930
|
[ARM repeat superfamily protein] |
0.318 |
-0.243 |
| 54 |
257144_at |
AT3G27300
|
G6PD5, glucose-6-phosphate dehydrogenase 5 |
0.317 |
0.106 |
| 55 |
248009_at |
AT5G56280
|
CSN6A, COP9 signalosome subunit 6A |
0.317 |
0.021 |
| 56 |
253520_at |
AT4G31410
|
unknown |
0.317 |
0.004 |
| 57 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
0.316 |
-0.229 |
| 58 |
251581_at |
AT3G58560
|
[DNAse I-like superfamily protein] |
0.315 |
-0.043 |
| 59 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
0.314 |
-0.365 |
| 60 |
261062_at |
AT1G07530
|
ATGRAS2, ARABIDOPSIS THALIANA GRAS (GAI, RGA, SCR) 2, GRAS2, GRAS (GAI, RGA, SCR) 2, SCL14, SCARECROW-like 14 |
0.314 |
-0.128 |
| 61 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.311 |
0.253 |
| 62 |
256856_at |
AT3G15110
|
unknown |
0.310 |
-0.004 |
| 63 |
252188_at |
AT3G50860
|
[Clathrin adaptor complex small chain family protein] |
0.309 |
0.022 |
| 64 |
255559_at |
AT4G02010
|
[Protein kinase superfamily protein] |
0.308 |
-0.230 |
| 65 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
0.307 |
-0.162 |
| 66 |
267339_at |
AT2G39870
|
unknown |
0.307 |
-0.233 |
| 67 |
262295_at |
AT1G27650
|
ATU2AF35A |
0.307 |
-0.005 |
| 68 |
261508_at |
AT1G71730
|
unknown |
0.305 |
-0.001 |
| 69 |
245502_at |
AT4G15640
|
unknown |
0.304 |
-0.041 |
| 70 |
261041_at |
AT1G17440
|
CKH1, CYTOKININ-HYPERSENSITIVE 1, EER4, ENHANCED ETHYLENE RESPONSE 4, TAF12B, TBP-ASSOCIATED FACTOR 12B |
0.301 |
0.039 |
| 71 |
265741_at |
AT2G01320
|
ABCG7, ATP-binding cassette G7 |
0.300 |
-0.155 |
| 72 |
253989_at |
AT4G26130
|
unknown |
0.298 |
0.190 |
| 73 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.298 |
0.535 |
| 74 |
250960_at |
AT5G02940
|
unknown |
0.297 |
0.256 |
| 75 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
0.296 |
-0.200 |
| 76 |
251849_at |
AT3G54610
|
BGT, BIG TOP, GCN5, general control non-repressible 5, HAC3, HAG01, HAG1, histone acetyltransferase of the GNAT family 1, HAT1, HISTONE ACETYLTRANSFERASE 1 |
0.296 |
-0.120 |
| 77 |
257530_at |
AT3G03040
|
[F-box/RNI-like superfamily protein] |
0.296 |
-0.093 |
| 78 |
262005_at |
AT1G64550
|
ABCF3, ATP-binding cassette F3, AtABCF3, AtGCN20, ATGCN3, general control non-repressible 3, GCN20, general control non-repressible 20, GCN3, general control non-repressible 3, SCORD5, susceptible to coronatine-deficient Pst DC3000 5 |
0.288 |
-0.105 |
| 79 |
247564_at |
AT5G61140
|
[U5 small nuclear ribonucleoprotein helicase] |
0.288 |
-0.063 |
| 80 |
259140_at |
AT3G10230
|
AtLCY, LYC, lycopene cyclase |
0.287 |
-0.291 |
| 81 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.287 |
-0.287 |
| 82 |
246087_at |
AT5G20580
|
unknown |
0.287 |
-0.106 |
| 83 |
249137_at |
AT5G43140
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.287 |
-0.084 |
| 84 |
260595_at |
AT1G55890
|
Tetratricopeptide repeat (TPR)-like superfamily protein |
0.285 |
-0.082 |
| 85 |
263688_at |
AT1G26920
|
unknown |
0.285 |
0.164 |
| 86 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
0.285 |
0.046 |
| 87 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.285 |
0.187 |
| 88 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.284 |
-0.069 |
| 89 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.284 |
-0.585 |
| 90 |
261398_at |
AT1G79610
|
ATNHX6, ARABIDOPSIS NA+/H+ ANTIPORTER 6, NHX6, Na+/H+ antiporter 6 |
0.284 |
0.103 |
| 91 |
264095_at |
AT1G79230
|
ATMST1, ATRDH1, ARABIDOPSIS THALIANA RHODANESE HOMOLOGUE 1, MST1, mercaptopyruvate sulfurtransferase 1, ST1, SULFURTRANSFERASE 1, STR1 |
0.283 |
-0.084 |
| 92 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
0.279 |
-0.282 |
| 93 |
251967_at |
AT3G53390
|
[Transducin/WD40 repeat-like superfamily protein] |
0.279 |
-0.009 |
| 94 |
246849_at |
AT5G26850
|
[Uncharacterized protein] |
0.278 |
-0.203 |
| 95 |
266450_s_at |
AT2G43190
|
POP4, similar to yeast POP4 |
0.278 |
0.017 |
| 96 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.275 |
-0.117 |
| 97 |
253283_at |
AT4G34090
|
unknown |
0.273 |
-0.314 |
| 98 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.273 |
-0.118 |
| 99 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.273 |
0.091 |
| 100 |
258804_at |
AT3G04760
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.271 |
-0.179 |
| 101 |
262348_at |
AT2G48160
|
[Tudor/PWWP/MBT domain-containing protein] |
0.271 |
-0.093 |
| 102 |
247036_at |
AT5G67130
|
[PLC-like phosphodiesterases superfamily protein] |
0.271 |
-0.042 |
| 103 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.270 |
-0.072 |
| 104 |
252313_at |
AT3G49390
|
CID10, CTC-interacting domain 10 |
0.270 |
0.111 |
| 105 |
258482_at |
AT3G02530
|
[TCP-1/cpn60 chaperonin family protein] |
0.270 |
-0.038 |
| 106 |
263212_at |
AT1G10510
|
emb2004, embryo defective 2004 |
0.269 |
-0.149 |
| 107 |
252192_at |
AT3G50000
|
ATCKA2, CKA2, casein kinase II, alpha chain 2 |
0.268 |
0.035 |
| 108 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
0.268 |
-0.146 |
| 109 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.268 |
0.384 |
| 110 |
252001_at |
AT3G52750
|
FTSZ2-2 |
0.268 |
-0.299 |
| 111 |
261577_at |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.267 |
-0.157 |
| 112 |
255570_at |
AT4G01100
|
ADNT1, adenine nucleotide transporter 1 |
0.266 |
-0.035 |
| 113 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.266 |
0.315 |
| 114 |
261522_at |
AT1G71710
|
[DNAse I-like superfamily protein] |
0.265 |
-0.210 |
| 115 |
261885_at |
AT1G80930
|
[MIF4G domain-containing protein / MA3 domain-containing protein] |
0.265 |
-0.024 |
| 116 |
262246_at |
AT1G48410
|
AGO1, ARGONAUTE 1, AtAGO1, ICU9 |
0.263 |
-0.123 |
| 117 |
254966_at |
AT4G11110
|
SPA2, SPA1-related 2 |
0.262 |
-0.115 |
| 118 |
253335_at |
AT4G33500
|
[Protein phosphatase 2C family protein] |
0.261 |
-0.217 |
| 119 |
252379_at |
AT3G47730
|
ABCA2, ATP-binding cassette A2, ATATH1, A. THALIANA ABC2 HOMOLOG 1, ATH1, ABC2 homolog 1 |
0.261 |
-0.019 |
| 120 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.260 |
0.047 |
| 121 |
251805_at |
AT3G55530
|
AtSDIR1, SDIR1, SALT- AND DROUGHT-INDUCED RING FINGER1 |
0.259 |
-0.016 |
| 122 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.258 |
-0.501 |
| 123 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
0.258 |
-0.250 |
| 124 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.257 |
0.029 |
| 125 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.257 |
0.495 |
| 126 |
248557_at |
AT5G49980
|
auxin F-box protein 5 |
0.256 |
-0.117 |
| 127 |
257655_at |
AT3G13350
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.256 |
-0.018 |
| 128 |
267513_at |
AT2G45620
|
URT1, UTP:RNA uridylyltransferase 1 |
0.256 |
-0.062 |
| 129 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.255 |
0.233 |
| 130 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
0.255 |
-0.085 |
| 131 |
245716_at |
AT5G08740
|
NDC1, NAD(P)H dehydrogenase C1 |
0.255 |
-0.046 |
| 132 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
0.254 |
-0.069 |
| 133 |
257913_at |
AT3G25540
|
LOH1, LAG One Homologue 1 |
0.254 |
-0.000 |
| 134 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.253 |
-0.173 |
| 135 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.252 |
0.133 |
| 136 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.252 |
0.048 |
| 137 |
263666_at |
AT1G04510
|
MAC3A, MOS4-associated complex 3A |
0.252 |
-0.062 |
| 138 |
264456_at |
AT1G10390
|
[Nucleoporin autopeptidase] |
0.252 |
-0.059 |
| 139 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
0.251 |
-0.180 |
| 140 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.251 |
-0.561 |
| 141 |
261376_at |
AT1G18660
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.250 |
-0.169 |
| 142 |
258963_at |
AT3G10550
|
AtMTM1, Arabidopsis thaliana myotubularin 1, MTM1, myotubularin 1 |
0.250 |
0.033 |
| 143 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.248 |
-0.254 |
| 144 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.247 |
-0.310 |
| 145 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.246 |
-0.025 |
| 146 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.246 |
0.378 |
| 147 |
247770_at |
AT5G58930
|
unknown |
0.245 |
-0.252 |
| 148 |
248957_at |
AT5G45620
|
[Proteasome component (PCI) domain protein] |
0.244 |
0.020 |
| 149 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
0.243 |
-0.021 |
| 150 |
263662_at |
AT1G04430
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.243 |
-0.138 |
| 151 |
258515_at |
AT3G06650
|
ACLB-1, ATP-citrate lyase B-1 |
0.243 |
-0.121 |
| 152 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
0.243 |
-0.255 |
| 153 |
267367_at |
AT2G44210
|
unknown |
0.242 |
-0.195 |
| 154 |
264928_at |
AT1G60710
|
ATB2 |
0.241 |
-0.039 |
| 155 |
265718_at |
AT2G03340
|
WRKY3, WRKY DNA-binding protein 3 |
0.241 |
-0.101 |
| 156 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.240 |
0.332 |
| 157 |
259378_at |
AT3G16310
|
1 |
0.239 |
0.049 |
| 158 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.239 |
-0.084 |
| 159 |
266939_at |
AT2G18960
|
AHA1, H(+)-ATPase 1, HA1, H(+)-ATPase 1, OST2, OPEN STOMATA 2, PMA, PLASMA MEMBRANE PROTON ATPASE |
0.239 |
-0.048 |
| 160 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
0.238 |
0.158 |
| 161 |
267520_at |
AT2G30460
|
[Nucleotide/sugar transporter family protein] |
0.237 |
-0.124 |
| 162 |
253300_at |
AT4G33580
|
ATBCA5, A. THALIANA BETA CARBONIC ANHYDRASE 5, BCA5, beta carbonic anhydrase 5 |
0.236 |
-0.229 |
| 163 |
266509_at |
AT2G47940
|
DEG2, degradation of periplasmic proteins 2, DEGP2, DEGP protease 2, EMB3117, EMBRYO DEFECTIVE 3117 |
0.235 |
-0.173 |
| 164 |
254615_at |
AT4G19210
|
ABCE2, ATP-binding cassette E2, ATRLI2, ARABIDOPSIS THALIANA RNASE L INHIBITOR PROTEIN 2, RLI2, RNAse l inhibitor protein 2 |
0.235 |
-0.039 |
| 165 |
254045_at |
AT4G25880
|
APUM6, pumilio 6, PUM6, pumilio 6 |
0.235 |
-0.153 |
| 166 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.234 |
-0.305 |
| 167 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.234 |
-0.114 |
| 168 |
264980_at |
AT1G27190
|
[Leucine-rich repeat protein kinase family protein] |
0.233 |
-0.210 |
| 169 |
259159_at |
AT3G05420
|
ACBP4, acyl-CoA binding protein 4, AtACBP4 |
0.232 |
-0.012 |
| 170 |
257702_at |
AT3G12670
|
emb2742, embryo defective 2742 |
0.231 |
-0.049 |
| 171 |
254048_at |
AT4G25680
|
[PPPDE putative thiol peptidase family protein] |
0.231 |
0.052 |
| 172 |
267430_at |
AT2G34860
|
EDA3, embryo sac development arrest 3 |
0.231 |
-0.043 |
| 173 |
264254_at |
AT1G09150
|
[pseudouridine synthase and archaeosine transglycosylase (PUA) domain-containing protein] |
0.231 |
0.031 |
| 174 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.230 |
0.293 |
| 175 |
266331_at |
AT2G01570
|
RGA, REPRESSOR OF GA, RGA1, REPRESSOR OF GA1-3 1 |
0.230 |
-0.203 |
| 176 |
247124_at |
AT5G66060
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.230 |
0.061 |
| 177 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
0.229 |
0.004 |
| 178 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.229 |
-0.348 |
| 179 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
0.229 |
-0.104 |
| 180 |
252075_at |
AT3G51730
|
[saposin B domain-containing protein] |
0.228 |
0.007 |
| 181 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
0.228 |
-0.092 |
| 182 |
265201_at |
AT2G36810
|
[ARM repeat superfamily protein] |
0.228 |
-0.157 |
| 183 |
254128_at |
AT4G24560
|
UBP16, ubiquitin-specific protease 16 |
0.228 |
-0.058 |
| 184 |
266523_at |
AT2G16950
|
ATTRN1, TRANSPORTIN 1, TRN1, transportin 1 |
0.227 |
-0.106 |
| 185 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.227 |
0.000 |
| 186 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
0.227 |
-0.230 |
| 187 |
246772_at |
AT5G27490
|
[Integral membrane Yip1 family protein] |
0.227 |
0.044 |
| 188 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.226 |
-0.139 |
| 189 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.226 |
0.046 |
| 190 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.226 |
-0.199 |
| 191 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.225 |
-0.379 |
| 192 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
0.225 |
-0.097 |
| 193 |
257981_at |
AT3G20770
|
AtEIN3, EIN3, ETHYLENE-INSENSITIVE3 |
0.225 |
0.065 |
| 194 |
258877_at |
AT3G03220
|
ATEXP13, EXPANSIN 13, ATEXPA13, expansin A13, ATHEXP ALPHA 1.22, EXP13, EXPA13, expansin A13 |
0.225 |
-0.136 |
| 195 |
262313_at |
AT1G70900
|
unknown |
0.223 |
-0.152 |
| 196 |
262856_at |
AT1G14910
|
[ENTH/ANTH/VHS superfamily protein] |
0.223 |
-0.055 |
| 197 |
253739_at |
AT4G28760
|
TRM20, TON1 Recruiting Motif 20 |
0.223 |
-0.315 |
| 198 |
254054_at |
AT4G25320
|
AHL3, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 |
0.222 |
0.022 |
| 199 |
264526_at |
AT1G10130
|
ATECA3, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 3, ECA3, endoplasmic reticulum-type calcium-transporting ATPase 3 |
0.222 |
-0.054 |
| 200 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
0.222 |
0.028 |
| 201 |
266753_at |
AT2G46990
|
IAA20, indole-3-acetic acid inducible 20 |
0.222 |
-0.071 |
| 202 |
253970_at |
AT4G26510
|
UKL4, uridine kinase-like 4 |
0.222 |
0.097 |
| 203 |
260496_at |
AT2G41700
|
ABCA1, ATP-binding cassette A1, AtABCA1, Arabidopsis thaliana ATP-binding cassette A1 |
0.221 |
-0.020 |
| 204 |
246141_at |
AT5G19920
|
[Transducin/WD40 repeat-like superfamily protein] |
0.220 |
-0.042 |
| 205 |
260626_at |
AT1G08040
|
unknown |
0.220 |
-0.017 |
| 206 |
263579_at |
AT2G17030
|
unknown |
0.220 |
0.034 |
| 207 |
247465_at |
AT5G62190
|
PRH75 |
0.220 |
-0.025 |
| 208 |
258684_at |
AT3G08680
|
[Leucine-rich repeat protein kinase family protein] |
0.219 |
-0.320 |
| 209 |
267163_at |
AT2G37520
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.219 |
0.157 |
| 210 |
259755_at |
AT1G71070
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.219 |
-0.031 |
| 211 |
245357_at |
AT4G17560
|
[Ribosomal protein L19 family protein] |
0.219 |
-0.132 |
| 212 |
261205_at |
AT1G12790
|
unknown |
0.219 |
0.136 |
| 213 |
256769_at |
AT3G13690
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.219 |
-0.268 |
| 214 |
254642_at |
AT4G18810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.218 |
-0.318 |
| 215 |
253210_at |
AT4G34840
|
ATMTAN2, ARABIDOPSIS METHYLTHIOADENOSINE NUCLEOSIDASE 2, ATMTN2, ARABIDOPSIS METHYLTHIOADENOSINE NUCLEOSIDASE 2, MTAN2, METHYLTHIOADENOSINE NUCLEOSIDASE 2, MTN2, methylthioadenosine nucleosidase 2 |
0.217 |
0.040 |
| 216 |
267374_at |
AT2G26230
|
[uricase / urate oxidase / nodulin 35, putative] |
0.217 |
0.163 |
| 217 |
263127_at |
AT1G78610
|
MSL6, mechanosensitive channel of small conductance-like 6 |
0.217 |
-0.187 |
| 218 |
250300_at |
AT5G11890
|
EMB3135, EMBRYO DEFECTIVE 3135 |
0.217 |
-0.160 |
| 219 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.217 |
-0.065 |
| 220 |
247558_at |
AT5G61060
|
ATHDA5, HDA05, histone deacetylase 5, HDA5 |
0.216 |
-0.057 |
| 221 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.216 |
-0.070 |
| 222 |
259542_at |
AT1G20575
|
DPMS1, dolichol phosphate mannose synthase 1 |
0.215 |
0.108 |
| 223 |
250178_at |
AT5G14430
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.215 |
-0.119 |
| 224 |
249370_at |
AT5G40710
|
[zinc finger (C2H2 type) family protein] |
0.215 |
0.011 |
| 225 |
267556_at |
AT2G32810
|
BGAL9, beta galactosidase 9, BGAL9, beta-galactosidase 9 |
0.214 |
-0.101 |
| 226 |
261269_at |
AT1G26690
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.214 |
0.242 |
| 227 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.213 |
-0.135 |
| 228 |
251260_at |
AT3G62130
|
AtLCD, LCD, L-cysteine desulfhydrase |
0.213 |
-0.069 |
| 229 |
256997_at |
AT3G14067
|
[Subtilase family protein] |
0.213 |
-0.064 |
| 230 |
247153_at |
AT5G65700
|
BAM1, BARELY ANY MERISTEM 1 |
0.213 |
-0.354 |
| 231 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
0.212 |
-0.288 |
| 232 |
262346_at |
AT1G63980
|
[D111/G-patch domain-containing protein] |
0.212 |
0.025 |
| 233 |
265387_at |
AT2G20670
|
unknown |
0.212 |
-0.242 |
| 234 |
256728_at |
AT3G25660
|
[Amidase family protein] |
0.211 |
-0.168 |
| 235 |
245625_at |
AT4G14160
|
[Sec23/Sec24 protein transport family protein] |
0.211 |
-0.045 |
| 236 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.211 |
0.212 |
| 237 |
255525_at |
AT4G02340
|
[alpha/beta-Hydrolases superfamily protein] |
0.210 |
0.288 |
| 238 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.210 |
-0.383 |
| 239 |
260596_at |
AT1G55900
|
EMB1860, embryo defective 1860, TIM50 |
0.210 |
-0.016 |
| 240 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.210 |
-0.127 |
| 241 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
0.210 |
-0.448 |
| 242 |
255586_at |
AT4G01560
|
MEE49, maternal effect embryo arrest 49 |
0.209 |
-0.006 |
| 243 |
260653_at |
AT1G32440
|
PKp3, plastidial pyruvate kinase 3 |
0.209 |
-0.216 |
| 244 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.209 |
0.195 |
| 245 |
263774_at |
AT2G40280
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.209 |
0.009 |
| 246 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
0.208 |
-0.031 |
| 247 |
245248_at |
AT4G15415
|
Protein phosphatase 2A regulatory B subunit family protein |
0.208 |
-0.110 |
| 248 |
249244_at |
AT5G42270
|
FTSH5, VAR1, VARIEGATED 1 |
0.208 |
-0.157 |
| 249 |
253000_at |
AT4G38360
|
LAZ1, LAZARUS 1 |
0.207 |
0.105 |
| 250 |
248451_at |
AT5G51180
|
[alpha/beta-Hydrolases superfamily protein] |
0.207 |
0.012 |
| 251 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
0.207 |
-0.125 |
| 252 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
0.207 |
0.194 |
| 253 |
246198_at |
AT4G36810
|
AtGGPPS11, AtLSU, GGPPS11, geranyl(geranyl)diphosphate synthase 11, GGPS1, geranylgeranyl pyrophosphate synthase 1, LSU, Large subunit of Heterodimeric geranyl(geranyl)diphosphate synthase |
0.207 |
-0.108 |
| 254 |
262800_at |
AT1G20960
|
emb1507, embryo defective 1507 |
0.207 |
-0.063 |
| 255 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
0.206 |
-0.557 |
| 256 |
261189_at |
AT1G33040
|
NACA5, nascent polypeptide-associated complex subunit alpha-like protein 5 |
0.206 |
-0.119 |
| 257 |
249216_at |
AT5G42240
|
scpl42, serine carboxypeptidase-like 42 |
0.206 |
-0.105 |
| 258 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
0.205 |
-0.086 |
| 259 |
259404_at |
AT1G17720
|
ATB BETA |
0.205 |
-0.072 |
| 260 |
248309_at |
AT5G52540
|
unknown |
0.204 |
-0.047 |
| 261 |
262963_at |
AT1G54220
|
[Dihydrolipoamide acetyltransferase, long form protein] |
0.204 |
-0.090 |
| 262 |
262506_at |
AT1G21640
|
ATNADK2, NAD KINASE 2, NADK2, NAD kinase 2 |
0.204 |
-0.115 |
| 263 |
256022_at |
AT1G58360
|
AAP1, amino acid permease 1, AtAAP1, NAT2, NEUTRAL AMINO ACID TRANSPORTER 2 |
0.203 |
0.115 |
| 264 |
255429_at |
AT4G03410
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.203 |
0.281 |
| 265 |
267605_at |
AT2G32920
|
ATPDI9, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 9, ATPDIL2-3, PDI-like 2-3, PDI9, PROTEIN DISULFIDE ISOMERASE 9, PDIL2-3, PDI-like 2-3 |
0.203 |
0.060 |
| 266 |
264286_at |
AT1G61870
|
PPR336, pentatricopeptide repeat 336 |
0.202 |
-0.066 |
| 267 |
251599_at |
AT3G57610
|
ADSS, adenylosuccinate synthase |
0.202 |
-0.071 |
| 268 |
264903_at |
AT1G23190
|
PGM3, phosphoglucomutase 3 |
0.201 |
-0.054 |
| 269 |
259819_at |
AT1G49820
|
ATMTK, S-methyl-5-thioribose kinase, MTK, S-methyl-5-thioribose kinase, MTK1, 5-methylthioribose kinase 1 |
0.201 |
0.057 |
| 270 |
258567_at |
AT3G04080
|
APY1, apyrase 1, ATAPY1, apyrase 1 |
0.201 |
0.044 |
| 271 |
258162_at |
AT3G17810
|
PYD1, pyrimidine 1 |
0.201 |
0.281 |
| 272 |
266422_at |
AT2G38650
|
GAUT7, galacturonosyltransferase 7, LGT7, LIKE GLYCOSYL TRANSFERASE 7 |
0.200 |
-0.121 |
| 273 |
254535_at |
AT4G19710
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH II, aspartate kinase-homoserine dehydrogenase ii |
0.200 |
-0.246 |
| 274 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.199 |
-0.148 |
| 275 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.199 |
-0.272 |
| 276 |
249748_at |
AT5G24620
|
[Pathogenesis-related thaumatin superfamily protein] |
0.199 |
0.082 |
| 277 |
250355_at |
AT5G11700
|
[LOCATED IN: vacuole] |
0.199 |
0.025 |
| 278 |
246157_at |
AT5G20080
|
[FAD/NAD(P)-binding oxidoreductase] |
0.198 |
0.049 |
| 279 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.198 |
-0.268 |
| 280 |
252649_at |
AT3G44680
|
AtHDA9, HDA09, HDA9, histone deacetylase 9 |
0.198 |
-0.020 |
| 281 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.198 |
-0.159 |
| 282 |
257756_at |
AT3G18680
|
[Amino acid kinase family protein] |
0.197 |
-0.067 |
| 283 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
0.197 |
0.119 |
| 284 |
252883_at |
AT4G39520
|
[GTP-binding protein-related] |
0.197 |
-0.041 |
| 285 |
265729_at |
AT2G31960
|
ATGSL03, glucan synthase-like 3, ATGSL3, GSL03, GSL03, glucan synthase-like 3 |
0.196 |
-0.104 |
| 286 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.196 |
0.758 |
| 287 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.195 |
-0.125 |
| 288 |
262176_at |
AT1G74960
|
ATKAS2, ARABIDOPSIS BETA-KETOACYL-ACP SYNTHETASE 2, FAB1, fatty acid biosynthesis 1, KAS2, BETA-KETOACYL-ACP SYNTHETASE 2 |
0.195 |
-0.149 |
| 289 |
262906_at |
AT1G59760
|
AtMTR4, MTR4, homolog of yeast MTR4 |
0.194 |
-0.100 |
| 290 |
267185_at |
AT2G43950
|
ATOEP37, ARABIDOPSIS CHLOROPLAST OUTER ENVELOPE PROTEIN 37, OEP37, chloroplast outer envelope protein 37 |
0.194 |
-0.128 |
| 291 |
248776_at |
AT5G47900
|
unknown |
0.194 |
-0.185 |
| 292 |
256401_at |
AT3G06200
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.194 |
-0.036 |
| 293 |
260928_at |
AT1G02720
|
AtGATL5, GATL5, galacturonosyltransferase-like 5 |
0.193 |
-0.086 |
| 294 |
257906_at |
AT3G25520
|
ATL5, ribosomal protein L5, OLI5, OLIGOCELLULA 5, PGY3, PIGGYBACK3, RPL5A, RIBOSOMAL PROTEIN L5 A |
0.193 |
-0.016 |
| 295 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
0.193 |
-0.829 |
| 296 |
257477_at |
AT1G10660
|
unknown |
0.193 |
-0.095 |
| 297 |
266819_at |
AT2G44870
|
unknown |
0.193 |
-0.041 |
| 298 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
0.193 |
-0.033 |
| 299 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.191 |
0.339 |
| 300 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.191 |
0.398 |
| 301 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.951 |
0.514 |
| 302 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-0.644 |
0.017 |
| 303 |
256499_at |
AT1G36640
|
unknown |
-0.627 |
0.084 |
| 304 |
246001_at |
AT5G20790
|
unknown |
-0.614 |
-0.646 |
| 305 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-0.603 |
0.298 |
| 306 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.599 |
0.866 |
| 307 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.570 |
0.422 |
| 308 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.569 |
-0.611 |
| 309 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
-0.556 |
0.115 |
| 310 |
247649_at |
AT5G60030
|
unknown |
-0.554 |
0.025 |
| 311 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.547 |
0.543 |
| 312 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.547 |
-0.207 |
| 313 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
-0.542 |
-0.089 |
| 314 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.534 |
0.212 |
| 315 |
253044_at |
AT4G37290
|
unknown |
-0.529 |
0.512 |
| 316 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.515 |
0.308 |
| 317 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-0.511 |
-0.177 |
| 318 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.506 |
-0.209 |
| 319 |
255128_at |
AT4G08310
|
unknown |
-0.491 |
-0.050 |
| 320 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
-0.489 |
0.267 |
| 321 |
253796_at |
AT4G28460
|
unknown |
-0.469 |
0.082 |
| 322 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.469 |
0.090 |
| 323 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
-0.467 |
0.004 |
| 324 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.461 |
-0.063 |
| 325 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
-0.461 |
0.095 |
| 326 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.457 |
0.162 |
| 327 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
-0.456 |
1.058 |
| 328 |
256569_at |
AT3G19550
|
unknown |
-0.450 |
0.577 |
| 329 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.445 |
-0.133 |
| 330 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
-0.443 |
0.682 |
| 331 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.440 |
0.410 |
| 332 |
250395_at |
AT5G10950
|
[Tudor/PWWP/MBT superfamily protein] |
-0.434 |
-0.055 |
| 333 |
246334_at |
AT3G44850
|
[Protein kinase superfamily protein] |
-0.434 |
0.064 |
| 334 |
249870_at |
AT5G23080
|
TGH, TOUGH |
-0.430 |
-0.080 |
| 335 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
-0.427 |
-0.051 |
| 336 |
252073_at |
AT3G51750
|
unknown |
-0.423 |
-0.474 |
| 337 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
-0.422 |
0.839 |
| 338 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.422 |
0.270 |
| 339 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.420 |
0.116 |
| 340 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.418 |
-0.238 |
| 341 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
-0.414 |
0.213 |
| 342 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.410 |
0.268 |
| 343 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.409 |
0.037 |
| 344 |
259385_at |
AT1G13470
|
unknown |
-0.408 |
0.479 |
| 345 |
264958_at |
AT1G76960
|
unknown |
-0.407 |
0.352 |
| 346 |
260307_at |
AT1G70620
|
[cyclin-related] |
-0.407 |
-0.056 |
| 347 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
-0.406 |
-0.243 |
| 348 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
-0.396 |
0.392 |
| 349 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
-0.393 |
0.316 |
| 350 |
245755_at |
AT1G35210
|
unknown |
-0.391 |
0.313 |
| 351 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
-0.389 |
0.023 |
| 352 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.387 |
-0.334 |
| 353 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.386 |
-0.016 |
| 354 |
265522_at |
AT2G06210
|
ELF8, EARLY FLOWERING 8, VIP6, VERNALIZATION INDEPENDENCE 6 |
-0.385 |
-0.083 |
| 355 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.385 |
0.399 |
| 356 |
249200_at |
AT5G42540
|
AtXRN2, EXORIBONUCLEASE 2, XRN2, exoribonuclease 2 |
-0.382 |
-0.134 |
| 357 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
-0.379 |
0.157 |
| 358 |
265024_at |
AT1G24600
|
unknown |
-0.378 |
0.569 |
| 359 |
251589_at |
AT3G58040
|
SINAT2, seven in absentia of Arabidopsis 2 |
-0.375 |
-0.062 |
| 360 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.373 |
0.902 |
| 361 |
246370_at |
AT1G51920
|
unknown |
-0.373 |
0.364 |
| 362 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.368 |
-0.380 |
| 363 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
-0.368 |
0.224 |
| 364 |
248379_at |
AT5G51700
|
ATRAR1, PBS2, PPHB SUSCEPTIBLE 2, RAR1, Required for Mla12 resistance 1, RPR2 |
-0.368 |
0.084 |
| 365 |
246321_at |
AT1G16640
|
[AP2/B3-like transcriptional factor family protein] |
-0.367 |
0.043 |
| 366 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
-0.365 |
0.064 |
| 367 |
260374_at |
AT1G73960
|
TAF2, TBP-associated factor 2 |
-0.364 |
-0.127 |
| 368 |
249740_at |
AT5G24500
|
unknown |
-0.361 |
0.021 |
| 369 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.361 |
-0.423 |
| 370 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.360 |
0.034 |
| 371 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
-0.358 |
0.043 |
| 372 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
-0.358 |
0.018 |
| 373 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.357 |
0.171 |
| 374 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.353 |
0.942 |
| 375 |
244901_at |
ATMG00640
|
ORF25 |
-0.352 |
0.044 |
| 376 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
-0.348 |
0.038 |
| 377 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.347 |
0.489 |
| 378 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.347 |
0.642 |
| 379 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.346 |
0.475 |
| 380 |
256777_at |
AT3G13780
|
[SMAD/FHA domain-containing protein ] |
-0.343 |
-0.042 |
| 381 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.342 |
-0.093 |
| 382 |
250065_at |
AT5G17910
|
unknown |
-0.341 |
0.084 |
| 383 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.341 |
-0.043 |
| 384 |
247253_at |
AT5G64790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.341 |
-0.006 |
| 385 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
-0.341 |
0.557 |
| 386 |
250796_at |
AT5G05300
|
unknown |
-0.340 |
0.524 |
| 387 |
256800_at |
AT3G20900
|
unknown |
-0.339 |
-0.059 |
| 388 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.339 |
0.371 |
| 389 |
250011_at |
AT5G18475
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.339 |
-0.026 |
| 390 |
263062_at |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.338 |
0.006 |
| 391 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
-0.335 |
0.499 |
| 392 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.334 |
0.852 |
| 393 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.332 |
-0.039 |
| 394 |
255063_at |
AT4G08970
|
[Mutator-like transposase family, has a 8.1e-28 P-value blast match to Q9SL18 /349-510 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.332 |
-0.003 |
| 395 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.332 |
0.489 |
| 396 |
245082_at |
AT2G23270
|
unknown |
-0.331 |
0.172 |
| 397 |
263798_at |
AT2G24640
|
UBP19, ubiquitin-specific protease 19 |
-0.331 |
-0.030 |
| 398 |
248702_at |
AT5G48420
|
unknown |
-0.326 |
0.000 |
| 399 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
-0.326 |
-0.027 |
| 400 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.326 |
-0.018 |
| 401 |
245143_at |
AT2G45450
|
ZPR1, LITTLE ZIPPER 1 |
-0.325 |
-0.197 |
| 402 |
258559_at |
AT3G06010
|
ATCHR12, CHR12, chromatin remodeling 12 |
-0.325 |
-0.117 |
| 403 |
266055_at |
AT2G40650
|
[PRP38 family protein] |
-0.324 |
-0.034 |
| 404 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.322 |
-0.070 |
| 405 |
253663_at |
AT4G30160
|
ATVLN4, VLN4, villin 4 |
-0.322 |
0.003 |
| 406 |
266283_at |
AT2G29210
|
[splicing factor PWI domain-containing protein] |
-0.322 |
-0.065 |
| 407 |
261239_at |
AT1G32930
|
AtGALT31A, GALT31A, glycosyltransferase of CAZY family GT31 A |
-0.321 |
-0.015 |
| 408 |
262391_at |
AT1G49530
|
GGPS6, geranylgeranyl pyrophosphate synthase 6 |
-0.320 |
0.217 |
| 409 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.318 |
-0.022 |
| 410 |
253859_at |
AT4G27657
|
unknown |
-0.317 |
0.474 |
| 411 |
267632_at |
AT2G42160
|
BRIZ1, BRAP2 RING ZnF UBP domain-containing protein 1 |
-0.315 |
0.119 |
| 412 |
267519_at |
AT2G30470
|
HSI2, high-level expression of sugar-inducible gene 2, VAL1, VP1/ABI3-LIKE 1 |
-0.315 |
-0.010 |
| 413 |
267226_at |
AT2G44010
|
unknown |
-0.314 |
0.209 |
| 414 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.314 |
0.288 |
| 415 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.313 |
-0.235 |
| 416 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.312 |
-0.188 |
| 417 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
-0.312 |
-0.157 |
| 418 |
248694_at |
AT5G48340
|
unknown |
-0.311 |
-0.091 |
| 419 |
255783_at |
AT1G19870
|
iqd32, IQ-domain 32 |
-0.311 |
-0.185 |
| 420 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.311 |
0.117 |
| 421 |
252494_at |
AT3G46760
|
LecRK-S.3, L-type lectin receptor kinase S.3 |
-0.309 |
-0.050 |
| 422 |
247141_at |
AT5G65560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.309 |
-0.003 |
| 423 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.308 |
0.382 |
| 424 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.308 |
-0.391 |
| 425 |
248988_at |
AT5G45190
|
[Cyclin family protein] |
-0.308 |
-0.042 |
| 426 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.307 |
-0.227 |
| 427 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.306 |
-0.229 |
| 428 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.306 |
0.254 |
| 429 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.304 |
0.833 |
| 430 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
-0.304 |
0.028 |
| 431 |
246841_at |
AT5G26700
|
[RmlC-like cupins superfamily protein] |
-0.302 |
0.011 |
| 432 |
266360_at |
AT2G32250
|
FRS2, FAR1-related sequence 2 |
-0.301 |
0.167 |
| 433 |
250310_at |
AT5G12230
|
MED19A |
-0.299 |
0.068 |
| 434 |
262156_at |
AT1G52680
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.298 |
-0.003 |
| 435 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.298 |
0.063 |
| 436 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.298 |
0.214 |
| 437 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.297 |
-0.340 |
| 438 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.296 |
-0.333 |
| 439 |
248616_at |
AT5G49580
|
[Chaperone DnaJ-domain superfamily protein] |
-0.296 |
-0.036 |
| 440 |
257331_at |
ATMG01330
|
ORF107H |
-0.295 |
-0.021 |
| 441 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.294 |
-0.284 |
| 442 |
263182_at |
AT1G05575
|
unknown |
-0.293 |
1.162 |
| 443 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.293 |
-0.040 |
| 444 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.293 |
-0.041 |
| 445 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.290 |
0.167 |
| 446 |
250098_at |
AT5G17350
|
unknown |
-0.290 |
0.193 |
| 447 |
252400_at |
AT3G48020
|
unknown |
-0.290 |
0.463 |
| 448 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.290 |
0.674 |
| 449 |
249936_at |
AT5G22450
|
unknown |
-0.289 |
-0.028 |
| 450 |
254200_at |
AT4G24110
|
unknown |
-0.289 |
0.395 |
| 451 |
266520_at |
AT2G23980
|
ATCNGC6, cyclic nucleotide-gated channel 6, CNGC6, cyclic nucleotide-gated channel 6 |
-0.288 |
-0.118 |
| 452 |
265077_at |
AT1G55530
|
[RING/U-box superfamily protein] |
-0.287 |
-0.141 |
| 453 |
247079_at |
AT5G66055
|
AKRP, ankyrin repeat protein, EMB16, EMBRYO DEFECTIVE 16, EMB2036, EMBRYO DEFECTIVE 2036 |
-0.287 |
-0.104 |
| 454 |
247708_at |
AT5G59550
|
unknown |
-0.286 |
0.352 |
| 455 |
246088_at |
AT5G20600
|
unknown |
-0.286 |
0.004 |
| 456 |
245821_at |
AT1G26270
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
-0.286 |
-0.140 |
| 457 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.286 |
-0.516 |
| 458 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
-0.285 |
0.038 |
| 459 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.285 |
0.215 |
| 460 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
-0.285 |
-0.146 |
| 461 |
249518_at |
AT5G38610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.284 |
-0.085 |
| 462 |
250774_at |
AT5G05450
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.284 |
-0.093 |
| 463 |
247448_at |
AT5G62770
|
unknown |
-0.283 |
0.052 |
| 464 |
248135_at |
AT5G54890
|
mCSF2, mitochondrial CAF-like splicing factor 2 |
-0.283 |
-0.089 |
| 465 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
-0.283 |
0.017 |
| 466 |
262812_at |
AT1G11690
|
unknown |
-0.282 |
-0.021 |
| 467 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.281 |
-0.035 |
| 468 |
247843_at |
AT5G58050
|
GDPDL6, Glycerophosphodiester phosphodiesterase (GDPD) like 6, SVL4, SHV3-like 4 |
-0.279 |
-0.014 |
| 469 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.278 |
-0.085 |
| 470 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
-0.277 |
0.780 |
| 471 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.277 |
0.220 |
| 472 |
260251_at |
AT1G74250
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.277 |
0.005 |
| 473 |
257537_at |
AT3G22350
|
[F-box and associated interaction domains-containing protein] |
-0.276 |
-0.020 |
| 474 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
-0.276 |
0.271 |
| 475 |
266661_at |
AT2G25820
|
ESE2, ethylene and salt inducible 2 |
-0.276 |
0.501 |
| 476 |
252023_at |
AT3G52920
|
unknown |
-0.275 |
-0.240 |
| 477 |
263526_at |
AT2G24830
|
[zinc finger (CCCH-type) family protein / D111/G-patch domain-containing protein] |
-0.274 |
0.007 |
| 478 |
259724_at |
AT1G60940
|
SNRK2-10, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-10, SNRK2.10, SNF1-related protein kinase 2.10, SRK2B, SNF1-RELATED KINASE 2B |
-0.274 |
-0.075 |
| 479 |
265762_at |
AT2G01240
|
[Reticulon family protein] |
-0.271 |
-0.001 |
| 480 |
245389_at |
AT4G17480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.270 |
0.083 |
| 481 |
248228_at |
AT5G53800
|
unknown |
-0.270 |
0.076 |
| 482 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
-0.270 |
-0.041 |
| 483 |
251010_at |
AT5G02550
|
unknown |
-0.269 |
0.135 |
| 484 |
257815_at |
AT3G25130
|
unknown |
-0.268 |
-0.281 |
| 485 |
263545_at |
AT2G21560
|
unknown |
-0.268 |
-0.365 |
| 486 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.267 |
0.545 |
| 487 |
260277_at |
AT1G80520
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.267 |
0.004 |
| 488 |
263972_at |
AT2G42760
|
unknown |
-0.267 |
0.451 |
| 489 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
-0.267 |
0.118 |
| 490 |
247394_at |
AT5G62865
|
unknown |
-0.265 |
0.344 |
| 491 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
-0.265 |
0.004 |
| 492 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.264 |
-0.253 |
| 493 |
244903_at |
ATMG00660
|
ORF149 |
-0.263 |
0.075 |
| 494 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.263 |
0.609 |
| 495 |
251633_at |
AT3G57460
|
[catalytics] |
-0.262 |
0.679 |
| 496 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.262 |
-0.036 |
| 497 |
247305_at |
AT5G63905
|
unknown |
-0.261 |
0.227 |
| 498 |
264629_at |
AT1G65540
|
AtLETM2, LETM2, leucine zipper-EF-hand-containing transmembrane protein 2 |
-0.261 |
0.084 |
| 499 |
259735_at |
AT1G64405
|
unknown |
-0.260 |
-0.028 |
| 500 |
258943_at |
AT3G10400
|
U11/U12-31K, U11/U12-31K |
-0.259 |
0.009 |
| 501 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.259 |
0.655 |
| 502 |
258354_at |
AT3G14320
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.259 |
-0.041 |
| 503 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
-0.258 |
0.080 |
| 504 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
-0.258 |
0.605 |
| 505 |
250179_at |
AT5G14440
|
[Surfeit locus protein 2 (SURF2)] |
-0.258 |
-0.054 |
| 506 |
249793_at |
AT5G23680
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.258 |
-0.087 |
| 507 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.258 |
-0.024 |
| 508 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.257 |
0.363 |
| 509 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.257 |
0.596 |
| 510 |
247913_at |
AT5G57510
|
unknown |
-0.257 |
0.239 |
| 511 |
250875_at |
AT5G04020
|
[calmodulin binding] |
-0.256 |
0.396 |
| 512 |
252532_at |
AT3G46570
|
[Glycosyl hydrolase superfamily protein] |
-0.256 |
-0.037 |
| 513 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
-0.256 |
0.328 |
| 514 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
-0.256 |
0.340 |
| 515 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
-0.256 |
-0.091 |
| 516 |
248601_at |
AT5G49400
|
[zinc knuckle (CCHC-type) family protein] |
-0.255 |
0.038 |
| 517 |
259718_at |
AT1G61040
|
VIP5, vernalization independence 5 |
-0.255 |
-0.039 |
| 518 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.255 |
0.073 |
| 519 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
-0.255 |
0.220 |
| 520 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
-0.254 |
-0.046 |
| 521 |
262453_at |
AT1G11240
|
unknown |
-0.254 |
0.073 |
| 522 |
261494_at |
AT1G28420
|
HB-1, homeobox-1, RLT1, RINGLET 1 |
-0.253 |
-0.069 |
| 523 |
256683_at |
AT3G52220
|
unknown |
-0.253 |
-0.003 |
| 524 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.253 |
0.008 |
| 525 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.253 |
0.716 |
| 526 |
263201_at |
AT1G05610
|
APS2, ADP-glucose pyrophosphorylase small subunit 2 |
-0.252 |
-0.029 |
| 527 |
248403_at |
AT5G51410
|
[LUC7 N_terminus domain-containing protein] |
-0.252 |
-0.016 |
| 528 |
263747_at |
AT2G21470
|
ATSAE2, SUMO-ACTIVATING ENZYME 2, EMB2764, EMBRYO DEFECTIVE 2764, SAE2, SUMO-activating enzyme 2 |
-0.251 |
0.110 |
| 529 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.250 |
-0.261 |
| 530 |
257067_at |
AT3G18270
|
pseudogene |
-0.250 |
0.006 |
| 531 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.250 |
-0.134 |
| 532 |
262093_at |
AT1G56145
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.250 |
0.100 |
| 533 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.250 |
0.629 |
| 534 |
250979_at |
AT5G03090
|
unknown |
-0.249 |
0.000 |
| 535 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.249 |
0.292 |
| 536 |
261987_at |
AT1G33710
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.248 |
-0.053 |
| 537 |
259190_at |
AT3G01780
|
TPLATE |
-0.248 |
-0.038 |
| 538 |
264865_at |
AT1G24120
|
ARL1, ARG1-like 1 |
-0.248 |
-0.088 |
| 539 |
262788_at |
AT1G10690
|
unknown |
-0.247 |
0.055 |
| 540 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.247 |
0.568 |
| 541 |
245588_at |
AT4G15030
|
unknown |
-0.247 |
-0.044 |
| 542 |
264082_at |
AT2G28570
|
unknown |
-0.247 |
0.038 |
| 543 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.247 |
0.048 |
| 544 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.246 |
0.022 |
| 545 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.246 |
0.669 |
| 546 |
254526_at |
AT4G19600
|
CYCT1;4 |
-0.246 |
-0.027 |
| 547 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.246 |
0.060 |
| 548 |
251049_at |
AT5G02430
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.246 |
-0.177 |
| 549 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.245 |
0.224 |
| 550 |
249086_at |
AT5G44180
|
RLT2, RINGLET 2 |
-0.245 |
-0.014 |
| 551 |
250638_at |
AT5G07540
|
ATGRP-6, ATGRP16, GRP16, glycine-rich protein 16 |
-0.244 |
-0.008 |
| 552 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
-0.244 |
0.291 |
| 553 |
246039_at |
AT5G19480
|
unknown |
-0.243 |
0.026 |
| 554 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.243 |
-0.232 |
| 555 |
250172_at |
AT5G14330
|
unknown |
-0.243 |
-0.020 |
| 556 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.243 |
-0.028 |
| 557 |
258203_at |
AT3G13950
|
unknown |
-0.242 |
0.665 |
| 558 |
267418_at |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
-0.242 |
0.194 |
| 559 |
259959_at |
AT1G53720
|
ATCYP59, CYCLOPHILIN 59, CYP59, cyclophilin 59 |
-0.242 |
-0.006 |
| 560 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.242 |
0.095 |
| 561 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
-0.241 |
0.058 |
| 562 |
258522_at |
AT3G06660
|
[PAPA-1-like family protein / zinc finger (HIT type) family protein] |
-0.241 |
-0.223 |
| 563 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.241 |
0.300 |
| 564 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.240 |
0.257 |
| 565 |
244923_s_at |
ATMG01020
|
ORF153B |
-0.240 |
0.060 |
| 566 |
245443_at |
AT4G16730
|
TPS02, terpene synthase 02 |
-0.240 |
0.883 |
| 567 |
253827_at |
AT4G28085
|
unknown |
-0.239 |
0.924 |
| 568 |
262783_at |
AT1G10850
|
[Leucine-rich repeat protein kinase family protein] |
-0.239 |
-0.481 |
| 569 |
266187_at |
AT2G38970
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.238 |
-0.369 |
| 570 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.238 |
0.229 |
| 571 |
267036_at |
AT2G38465
|
unknown |
-0.238 |
0.138 |
| 572 |
267087_at |
AT2G32460
|
ATM1, ARABIDOPSIS THALIANA MYB 1, ATMYB101, MYB101, myb domain protein 101 |
-0.237 |
-0.012 |
| 573 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.236 |
0.274 |
| 574 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
-0.235 |
-0.001 |
| 575 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.234 |
-0.018 |
| 576 |
267214_at |
AT2G43970
|
AtLARP6b, LARP6b, La related protein 6b |
-0.234 |
-0.012 |
| 577 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.234 |
-0.500 |
| 578 |
246946_at |
AT5G25070
|
unknown |
-0.234 |
0.014 |
| 579 |
260600_at |
AT1G55950
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.233 |
0.009 |
| 580 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
-0.233 |
0.528 |
| 581 |
261891_at |
AT1G80790
|
FDM5, factor of DNA methylation 5 |
-0.233 |
-0.171 |
| 582 |
265039_at |
AT1G04000
|
unknown |
-0.233 |
-0.188 |
| 583 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
-0.232 |
0.258 |
| 584 |
250273_at |
AT5G13010
|
EMB3011, embryo defective 3011 |
-0.232 |
-0.057 |
| 585 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.232 |
-0.189 |
| 586 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.232 |
-0.074 |
| 587 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.231 |
0.503 |
| 588 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.231 |
-0.040 |
| 589 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.230 |
-0.200 |
| 590 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.230 |
-0.104 |
| 591 |
256348_at |
AT1G54880
|
unknown |
-0.230 |
-0.038 |
| 592 |
250944_at |
AT5G03380
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.229 |
-0.057 |
| 593 |
245461_at |
AT4G17000
|
unknown |
-0.229 |
-0.082 |
| 594 |
250556_at |
AT5G07920
|
ATDGK1, DIACYLGLYCEROL KINASE 1, DGK1, diacylglycerol kinase1 |
-0.229 |
0.283 |
| 595 |
261503_at |
AT1G71691
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.229 |
-0.064 |
| 596 |
251487_at |
AT3G59760
|
ATCS-C, ARABIDOPSIS THALIANA CYSTEINSYNTHASE-C, OASC, O-acetylserine (thiol) lyase isoform C |
-0.228 |
0.124 |
| 597 |
259638_at |
AT1G52360
|
[Coatomer, beta' subunit] |
-0.228 |
-0.053 |
| 598 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.227 |
0.830 |
| 599 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
-0.227 |
-0.072 |
| 600 |
260883_at |
AT1G29270
|
unknown |
-0.227 |
-0.047 |