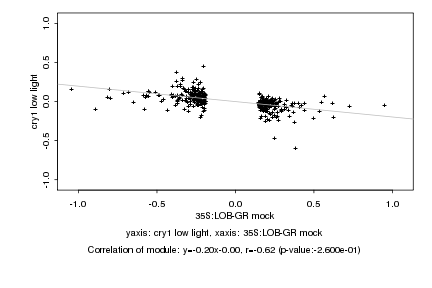
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:LOB-GR mock |
Log2 signal ratio
cry1 low light |
| 1 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.948 |
-0.046 |
| 2 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.725 |
-0.062 |
| 3 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.620 |
-0.203 |
| 4 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.617 |
-0.020 |
| 5 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.566 |
0.072 |
| 6 |
245532_at |
AT4G15110
|
CYP97B3, cytochrome P450, family 97, subfamily B, polypeptide 3 |
0.546 |
-0.002 |
| 7 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.532 |
-0.126 |
| 8 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.495 |
-0.211 |
| 9 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.438 |
-0.112 |
| 10 |
256603_at |
AT3G28270
|
unknown |
0.434 |
-0.019 |
| 11 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.418 |
-0.045 |
| 12 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.407 |
-0.076 |
| 13 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.400 |
-0.014 |
| 14 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.381 |
-0.592 |
| 15 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.374 |
-0.021 |
| 16 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.374 |
-0.266 |
| 17 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.367 |
-0.133 |
| 18 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.363 |
-0.017 |
| 19 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.359 |
-0.051 |
| 20 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.357 |
-0.050 |
| 21 |
246001_at |
AT5G20790
|
unknown |
0.340 |
-0.189 |
| 22 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.337 |
-0.110 |
| 23 |
262533_at |
AT1G17090
|
unknown |
0.325 |
0.015 |
| 24 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.320 |
-0.072 |
| 25 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.311 |
-0.058 |
| 26 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.310 |
-0.094 |
| 27 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.307 |
-0.028 |
| 28 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.292 |
-0.091 |
| 29 |
247800_at |
AT5G58570
|
unknown |
0.290 |
-0.117 |
| 30 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.290 |
-0.109 |
| 31 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.287 |
0.002 |
| 32 |
250696_at |
AT5G06790
|
unknown |
0.277 |
0.044 |
| 33 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.277 |
0.007 |
| 34 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.273 |
-0.003 |
| 35 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.272 |
-0.234 |
| 36 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.269 |
-0.055 |
| 37 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.263 |
-0.183 |
| 38 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.262 |
-0.133 |
| 39 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.259 |
-0.193 |
| 40 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.258 |
-0.025 |
| 41 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.257 |
-0.033 |
| 42 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.255 |
-0.016 |
| 43 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.251 |
-0.087 |
| 44 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.251 |
-0.054 |
| 45 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.250 |
0.031 |
| 46 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.247 |
-0.053 |
| 47 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.247 |
0.014 |
| 48 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
0.247 |
-0.100 |
| 49 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.246 |
-0.168 |
| 50 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.245 |
-0.471 |
| 51 |
262162_at |
AT1G78020
|
unknown |
0.241 |
-0.047 |
| 52 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
0.238 |
-0.015 |
| 53 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.235 |
0.007 |
| 54 |
251811_at |
AT3G54990
|
SMZ, SCHLAFMUTZE |
0.235 |
-0.013 |
| 55 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.234 |
-0.140 |
| 56 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.234 |
-0.008 |
| 57 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.233 |
0.045 |
| 58 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
0.231 |
-0.012 |
| 59 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.231 |
-0.200 |
| 60 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
0.229 |
-0.042 |
| 61 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.229 |
-0.055 |
| 62 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
0.227 |
-0.001 |
| 63 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.224 |
0.028 |
| 64 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.221 |
-0.054 |
| 65 |
254146_at |
AT4G24260
|
ATGH9A3, glycosyl hydrolase 9A3, GH9A3, glycosyl hydrolase 9A3, KOR3 |
0.221 |
-0.003 |
| 66 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.218 |
-0.165 |
| 67 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.218 |
0.020 |
| 68 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
0.217 |
0.003 |
| 69 |
249728_at |
AT5G24390
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.215 |
0.007 |
| 70 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.212 |
0.003 |
| 71 |
254022_at |
AT4G25750
|
ABCG4, ATP-binding cassette G4 |
0.212 |
-0.013 |
| 72 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.211 |
-0.141 |
| 73 |
264636_at |
AT1G65490
|
unknown |
0.211 |
-0.064 |
| 74 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.211 |
-0.230 |
| 75 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.210 |
0.006 |
| 76 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
0.208 |
0.068 |
| 77 |
253322_at |
AT4G33980
|
unknown |
0.206 |
-0.049 |
| 78 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.204 |
-0.227 |
| 79 |
265990_at |
AT2G24280
|
[alpha/beta-Hydrolases superfamily protein] |
0.204 |
0.007 |
| 80 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.202 |
-0.119 |
| 81 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.201 |
-0.001 |
| 82 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.201 |
-0.094 |
| 83 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.200 |
0.017 |
| 84 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.200 |
-0.001 |
| 85 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.198 |
-0.024 |
| 86 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.197 |
0.032 |
| 87 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.197 |
-0.117 |
| 88 |
249377_at |
AT5G40690
|
unknown |
0.193 |
-0.041 |
| 89 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.193 |
0.045 |
| 90 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
0.192 |
-0.061 |
| 91 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.192 |
-0.078 |
| 92 |
266738_at |
AT2G47010
|
unknown |
0.190 |
-0.051 |
| 93 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
0.190 |
0.014 |
| 94 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.188 |
-0.116 |
| 95 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.188 |
-0.011 |
| 96 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.188 |
-0.054 |
| 97 |
251907_at |
AT3G53760
|
ATGCP4, GCP4, GAMMA-TUBULIN COMPLEX PROTEIN 4 |
0.188 |
0.008 |
| 98 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
0.187 |
0.015 |
| 99 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.187 |
-0.186 |
| 100 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.187 |
-0.244 |
| 101 |
248335_at |
AT5G52450
|
[MATE efflux family protein] |
0.186 |
-0.066 |
| 102 |
253008_at |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
0.186 |
-0.004 |
| 103 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.186 |
-0.083 |
| 104 |
251292_at |
AT3G61920
|
unknown |
0.185 |
-0.007 |
| 105 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
0.184 |
0.017 |
| 106 |
249482_at |
AT5G38980
|
unknown |
0.183 |
-0.005 |
| 107 |
255676_at |
AT4G00490
|
BAM2, beta-amylase 2, BMY9, BETA-AMYLASE 9 |
0.183 |
-0.044 |
| 108 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.183 |
-0.045 |
| 109 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
0.182 |
-0.036 |
| 110 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.180 |
0.000 |
| 111 |
249759_at |
AT5G24380
|
ATYSL2, YELLOW STRIPE LIKE 2, YSL2, YELLOW STRIPE like 2 |
0.180 |
0.012 |
| 112 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.180 |
-0.092 |
| 113 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.179 |
-0.075 |
| 114 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.179 |
-0.092 |
| 115 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.177 |
0.016 |
| 116 |
247795_at |
AT5G58620
|
TZF9, tandem zinc finger protein 9 |
0.177 |
-0.011 |
| 117 |
263068_at |
AT2G17580
|
[Polynucleotide adenylyltransferase family protein] |
0.176 |
-0.002 |
| 118 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.176 |
-0.117 |
| 119 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
0.175 |
0.038 |
| 120 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.174 |
-0.035 |
| 121 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.174 |
0.032 |
| 122 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.173 |
-0.077 |
| 123 |
262811_at |
AT1G11700
|
unknown |
0.172 |
-0.072 |
| 124 |
257746_at |
AT3G29200
|
ATCM1, ARABIDOPSIS THALIANA CHORISMATE MUTASE 1, CM1, chorismate mutase 1 |
0.170 |
-0.037 |
| 125 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.169 |
-0.087 |
| 126 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.169 |
0.005 |
| 127 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.168 |
-0.083 |
| 128 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
0.168 |
-0.090 |
| 129 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.168 |
0.066 |
| 130 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.167 |
-0.080 |
| 131 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.167 |
0.054 |
| 132 |
265503_at |
AT2G15510
|
[non-LTR retrotransposon family (LINE), has a 3.0e-20 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
0.166 |
0.058 |
| 133 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.165 |
-0.184 |
| 134 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.165 |
-0.009 |
| 135 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.165 |
-0.077 |
| 136 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.164 |
-0.049 |
| 137 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.164 |
-0.033 |
| 138 |
249702_at |
AT5G35570
|
[O-fucosyltransferase family protein] |
0.164 |
-0.030 |
| 139 |
246247_at |
AT4G36640
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.163 |
-0.045 |
| 140 |
263134_at |
AT1G78570
|
ATRHM1, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 1, RHM1, rhamnose biosynthesis 1, ROL1, REPRESSOR OF LRX1 1 |
0.162 |
-0.065 |
| 141 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.161 |
-0.016 |
| 142 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.161 |
0.052 |
| 143 |
264232_at |
AT1G67470
|
[Protein kinase superfamily protein] |
0.161 |
-0.076 |
| 144 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
0.160 |
-0.021 |
| 145 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.158 |
-0.063 |
| 146 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.158 |
-0.021 |
| 147 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.158 |
-0.002 |
| 148 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
0.158 |
-0.015 |
| 149 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.158 |
-0.029 |
| 150 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
0.158 |
0.019 |
| 151 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.157 |
-0.213 |
| 152 |
254112_at |
AT4G24970
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.157 |
-0.050 |
| 153 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.157 |
0.002 |
| 154 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
0.157 |
0.022 |
| 155 |
253374_at |
AT4G33140
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.156 |
-0.063 |
| 156 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.156 |
-0.078 |
| 157 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.155 |
-0.108 |
| 158 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.154 |
-0.045 |
| 159 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
0.153 |
-0.033 |
| 160 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.152 |
-0.045 |
| 161 |
246225_at |
AT4G36910
|
CBSX1, CBS domain containing protein 1, CDCP2, CYSTATHIONE [BETA]-SYNTHASE DOMAIN-CONTAINING PROTEIN 2, LEJ2, LOSS OF THE TIMING OF ET AND JA BIOSYNTHESIS 2 |
0.152 |
-0.022 |
| 162 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.151 |
-0.042 |
| 163 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.151 |
-0.026 |
| 164 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
0.150 |
-0.025 |
| 165 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.149 |
-0.059 |
| 166 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.148 |
-0.046 |
| 167 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
0.148 |
-0.028 |
| 168 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
0.148 |
-0.068 |
| 169 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.147 |
0.091 |
| 170 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.147 |
0.106 |
| 171 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.147 |
-0.027 |
| 172 |
254723_at |
AT4G13510
|
AMT1;1, ammonium transporter 1;1, ATAMT1, ARABIDOPSIS THALIANA AMMONIUM TRANSPORT 1, ATAMT1;1 |
0.147 |
-0.030 |
| 173 |
259856_at |
AT1G68440
|
unknown |
0.146 |
-0.001 |
| 174 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.145 |
-0.020 |
| 175 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
0.145 |
-0.018 |
| 176 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-1.049 |
0.165 |
| 177 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.897 |
-0.101 |
| 178 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.818 |
0.062 |
| 179 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.804 |
0.157 |
| 180 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.801 |
0.047 |
| 181 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.714 |
0.110 |
| 182 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.683 |
0.120 |
| 183 |
246045_at |
AT5G19430
|
[RING/U-box superfamily protein] |
-0.652 |
-0.008 |
| 184 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.590 |
0.081 |
| 185 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.585 |
-0.096 |
| 186 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.576 |
0.064 |
| 187 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
-0.571 |
0.088 |
| 188 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.558 |
0.131 |
| 189 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.557 |
0.140 |
| 190 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.555 |
0.071 |
| 191 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.552 |
0.125 |
| 192 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.515 |
0.118 |
| 193 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.492 |
0.086 |
| 194 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.487 |
0.079 |
| 195 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.472 |
0.011 |
| 196 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.459 |
0.026 |
| 197 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.437 |
-0.111 |
| 198 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.412 |
0.101 |
| 199 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.410 |
0.086 |
| 200 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.405 |
0.204 |
| 201 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.402 |
0.064 |
| 202 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.396 |
0.085 |
| 203 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.388 |
-0.047 |
| 204 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.384 |
0.075 |
| 205 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.380 |
0.374 |
| 206 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.376 |
0.263 |
| 207 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.375 |
0.010 |
| 208 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.373 |
-0.016 |
| 209 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.373 |
0.196 |
| 210 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.371 |
0.018 |
| 211 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.370 |
0.089 |
| 212 |
247754_at |
AT5G59080
|
unknown |
-0.368 |
0.030 |
| 213 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.362 |
0.051 |
| 214 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.358 |
0.058 |
| 215 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.355 |
0.230 |
| 216 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.353 |
0.193 |
| 217 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.348 |
0.102 |
| 218 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.343 |
0.280 |
| 219 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.341 |
0.017 |
| 220 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.341 |
0.079 |
| 221 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.339 |
0.307 |
| 222 |
247903_at |
AT5G57340
|
unknown |
-0.335 |
0.169 |
| 223 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.335 |
0.176 |
| 224 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.329 |
-0.092 |
| 225 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.329 |
0.157 |
| 226 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.323 |
0.140 |
| 227 |
254954_at |
AT4G10910
|
unknown |
-0.322 |
0.019 |
| 228 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.317 |
0.076 |
| 229 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.316 |
0.003 |
| 230 |
264774_at |
AT1G22890
|
unknown |
-0.314 |
0.141 |
| 231 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
-0.313 |
0.042 |
| 232 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.309 |
-0.008 |
| 233 |
260668_at |
AT1G19530
|
unknown |
-0.305 |
-0.124 |
| 234 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.305 |
-0.061 |
| 235 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.304 |
0.116 |
| 236 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.303 |
0.160 |
| 237 |
247025_at |
AT5G67030
|
ABA1, ABA DEFICIENT 1, ATABA1, ARABIDOPSIS THALIANA ABA DEFICIENT 1, ATZEP, ARABIDOPSIS THALIANA ZEAXANTHIN EPOXIDASE, IBS3, IMPAIRED IN BABA-INDUCED STERILITY 3, LOS6, LOW EXPRESSION OF OSMOTIC STRESS-RESPONSIVE GENES 6, NPQ2, NON-PHOTOCHEMICAL QUENCHING 2, ZEP, ZEAXANTHIN EPOXIDASE |
-0.302 |
0.045 |
| 238 |
259275_at |
AT3G01060
|
unknown |
-0.297 |
0.039 |
| 239 |
246487_at |
AT5G16030
|
unknown |
-0.292 |
0.041 |
| 240 |
262236_at |
AT1G48330
|
unknown |
-0.292 |
-0.031 |
| 241 |
264314_at |
AT1G70420
|
unknown |
-0.290 |
0.094 |
| 242 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.289 |
0.118 |
| 243 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.288 |
0.007 |
| 244 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.288 |
0.054 |
| 245 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.285 |
0.079 |
| 246 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.284 |
0.110 |
| 247 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.276 |
0.069 |
| 248 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.276 |
0.182 |
| 249 |
259373_at |
AT1G69160
|
unknown |
-0.275 |
0.163 |
| 250 |
258196_at |
AT3G13980
|
unknown |
-0.274 |
0.114 |
| 251 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.273 |
-0.052 |
| 252 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.272 |
0.182 |
| 253 |
265387_at |
AT2G20670
|
unknown |
-0.272 |
0.022 |
| 254 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.272 |
0.096 |
| 255 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.272 |
0.035 |
| 256 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.271 |
0.246 |
| 257 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.267 |
0.149 |
| 258 |
260956_at |
AT1G06040
|
BBX24, B-box domain protein 24, STO, SALT TOLERANCE |
-0.267 |
0.064 |
| 259 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
-0.266 |
0.096 |
| 260 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.266 |
0.031 |
| 261 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.265 |
0.041 |
| 262 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.263 |
0.029 |
| 263 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.263 |
0.140 |
| 264 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.261 |
-0.008 |
| 265 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-0.260 |
0.094 |
| 266 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.260 |
0.170 |
| 267 |
247899_at |
AT5G57345
|
unknown |
-0.258 |
0.034 |
| 268 |
258939_at |
AT3G10020
|
unknown |
-0.256 |
0.049 |
| 269 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.255 |
0.034 |
| 270 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.255 |
0.138 |
| 271 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.250 |
0.288 |
| 272 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.249 |
0.048 |
| 273 |
259015_at |
AT3G07350
|
unknown |
-0.249 |
0.045 |
| 274 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.249 |
0.100 |
| 275 |
261772_at |
AT1G76240
|
unknown |
-0.248 |
0.025 |
| 276 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.248 |
0.003 |
| 277 |
247295_at |
AT5G64180
|
unknown |
-0.245 |
0.059 |
| 278 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.245 |
-0.036 |
| 279 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.244 |
-0.021 |
| 280 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.244 |
0.145 |
| 281 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.243 |
-0.042 |
| 282 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.241 |
0.072 |
| 283 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.241 |
0.086 |
| 284 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.240 |
-0.001 |
| 285 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.240 |
0.178 |
| 286 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.239 |
0.040 |
| 287 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.238 |
0.226 |
| 288 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.238 |
0.026 |
| 289 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.238 |
0.108 |
| 290 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.238 |
0.109 |
| 291 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
-0.238 |
-0.041 |
| 292 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.237 |
0.001 |
| 293 |
249134_at |
AT5G43150
|
unknown |
-0.236 |
0.053 |
| 294 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.235 |
0.031 |
| 295 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.234 |
0.044 |
| 296 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.233 |
0.104 |
| 297 |
263607_at |
AT2G16270
|
unknown |
-0.233 |
0.030 |
| 298 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.232 |
0.065 |
| 299 |
259982_at |
AT1G76410
|
ATL8 |
-0.231 |
0.041 |
| 300 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.231 |
0.125 |
| 301 |
246997_at |
AT5G67390
|
unknown |
-0.229 |
0.027 |
| 302 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.228 |
-0.026 |
| 303 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
-0.228 |
0.086 |
| 304 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.227 |
0.256 |
| 305 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.225 |
-0.201 |
| 306 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.225 |
0.078 |
| 307 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.225 |
0.062 |
| 308 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.223 |
-0.004 |
| 309 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.223 |
0.081 |
| 310 |
261247_at |
AT1G20070
|
unknown |
-0.222 |
0.067 |
| 311 |
261567_at |
AT1G33055
|
unknown |
-0.222 |
-0.175 |
| 312 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.221 |
-0.028 |
| 313 |
250975_at |
AT5G03050
|
unknown |
-0.219 |
0.012 |
| 314 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.218 |
0.129 |
| 315 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.217 |
0.025 |
| 316 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.216 |
0.135 |
| 317 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.215 |
0.010 |
| 318 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
-0.214 |
-0.067 |
| 319 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.213 |
0.106 |
| 320 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.213 |
-0.023 |
| 321 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.212 |
0.002 |
| 322 |
247585_at |
AT5G60680
|
unknown |
-0.210 |
0.071 |
| 323 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.210 |
0.158 |
| 324 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.209 |
0.176 |
| 325 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.209 |
0.008 |
| 326 |
261026_at |
AT1G01240
|
unknown |
-0.208 |
0.102 |
| 327 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.207 |
0.081 |
| 328 |
262719_at |
AT1G43590
|
unknown |
-0.205 |
0.459 |
| 329 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.204 |
0.095 |
| 330 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.204 |
-0.118 |
| 331 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.203 |
0.058 |
| 332 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.202 |
0.063 |
| 333 |
264379_at |
AT2G25200
|
unknown |
-0.202 |
0.001 |
| 334 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
-0.201 |
-0.008 |
| 335 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.201 |
0.098 |
| 336 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.201 |
-0.110 |
| 337 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.200 |
0.036 |
| 338 |
249355_at |
AT5G40500
|
unknown |
-0.200 |
0.019 |
| 339 |
258468_at |
AT3G06070
|
unknown |
-0.200 |
0.112 |
| 340 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.198 |
-0.080 |
| 341 |
252412_at |
AT3G47295
|
unknown |
-0.198 |
0.077 |
| 342 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.198 |
-0.029 |
| 343 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.197 |
0.040 |
| 344 |
263118_at |
AT1G03090
|
MCCA |
-0.196 |
-0.019 |
| 345 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.195 |
0.093 |
| 346 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.195 |
0.007 |
| 347 |
260877_at |
AT1G21500
|
unknown |
-0.195 |
0.078 |
| 348 |
266125_at |
AT2G45050
|
GATA2, GATA transcription factor 2 |
-0.194 |
0.001 |
| 349 |
250656_at |
AT5G06970
|
unknown |
-0.193 |
0.003 |