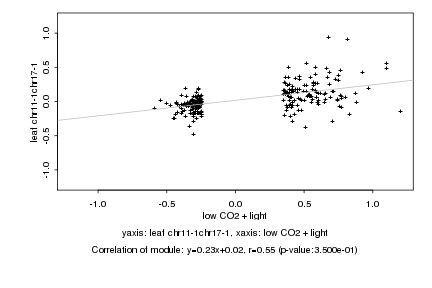
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
low CO2 + light |
Log2 signal ratio
leaf chr11-1chr17-1 |
| 1 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.199 |
-0.134 |
| 2 |
264580_at |
AT1G05340
|
unknown |
1.096 |
0.570 |
| 3 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.096 |
0.497 |
| 4 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.963 |
0.196 |
| 5 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.918 |
0.436 |
| 6 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.880 |
-0.004 |
| 7 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.872 |
0.119 |
| 8 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.828 |
-0.189 |
| 9 |
252539_at |
AT3G45730
|
unknown |
0.813 |
0.918 |
| 10 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.797 |
0.064 |
| 11 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.768 |
-0.081 |
| 12 |
264774_at |
AT1G22890
|
unknown |
0.766 |
0.052 |
| 13 |
259479_at |
AT1G19020
|
unknown |
0.765 |
0.077 |
| 14 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.762 |
0.465 |
| 15 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.759 |
0.096 |
| 16 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.756 |
-0.066 |
| 17 |
253827_at |
AT4G28085
|
unknown |
0.749 |
0.392 |
| 18 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.747 |
0.321 |
| 19 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.742 |
0.042 |
| 20 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.742 |
0.023 |
| 21 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.725 |
0.332 |
| 22 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.707 |
0.159 |
| 23 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.706 |
0.148 |
| 24 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.705 |
-0.291 |
| 25 |
247835_at |
AT5G57910
|
unknown |
0.686 |
0.068 |
| 26 |
263475_at |
AT2G31945
|
unknown |
0.684 |
0.252 |
| 27 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.678 |
0.431 |
| 28 |
249454_at |
AT5G39520
|
unknown |
0.676 |
0.945 |
| 29 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.669 |
0.355 |
| 30 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.658 |
0.485 |
| 31 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.654 |
0.130 |
| 32 |
259979_at |
AT1G76600
|
unknown |
0.649 |
0.038 |
| 33 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.647 |
-0.009 |
| 34 |
263182_at |
AT1G05575
|
unknown |
0.645 |
0.108 |
| 35 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.617 |
0.131 |
| 36 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.601 |
-0.032 |
| 37 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
0.600 |
0.026 |
| 38 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.598 |
0.128 |
| 39 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.595 |
-0.010 |
| 40 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.594 |
0.276 |
| 41 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.588 |
0.060 |
| 42 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.586 |
0.065 |
| 43 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.585 |
0.144 |
| 44 |
258275_at |
AT3G15760
|
unknown |
0.582 |
0.175 |
| 45 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.582 |
0.268 |
| 46 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.581 |
0.409 |
| 47 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.577 |
0.507 |
| 48 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.566 |
0.241 |
| 49 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.566 |
0.290 |
| 50 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.563 |
0.273 |
| 51 |
263881_at |
AT2G21820
|
unknown |
0.561 |
0.031 |
| 52 |
266259_at |
AT2G27830
|
unknown |
0.557 |
0.180 |
| 53 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.548 |
0.086 |
| 54 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.547 |
-0.001 |
| 55 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
0.545 |
0.362 |
| 56 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.541 |
0.077 |
| 57 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.537 |
0.108 |
| 58 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.535 |
0.078 |
| 59 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.529 |
0.097 |
| 60 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.529 |
0.098 |
| 61 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.519 |
0.085 |
| 62 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.517 |
0.236 |
| 63 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.515 |
0.563 |
| 64 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.512 |
0.150 |
| 65 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.510 |
-0.366 |
| 66 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.498 |
0.245 |
| 67 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.497 |
0.026 |
| 68 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.495 |
0.148 |
| 69 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.479 |
-0.127 |
| 70 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.474 |
0.015 |
| 71 |
262801_at |
AT1G21010
|
unknown |
0.472 |
0.031 |
| 72 |
248432_at |
AT5G51390
|
unknown |
0.465 |
0.324 |
| 73 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
0.460 |
0.375 |
| 74 |
263632_at |
AT2G04795
|
unknown |
0.460 |
0.181 |
| 75 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.458 |
-0.120 |
| 76 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.457 |
0.055 |
| 77 |
261567_at |
AT1G33055
|
unknown |
0.449 |
-0.046 |
| 78 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.447 |
0.133 |
| 79 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.446 |
0.180 |
| 80 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.437 |
0.344 |
| 81 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
0.433 |
0.187 |
| 82 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.426 |
0.024 |
| 83 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.425 |
-0.185 |
| 84 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.422 |
0.154 |
| 85 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
0.417 |
0.155 |
| 86 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.414 |
-0.002 |
| 87 |
260544_at |
AT2G43540
|
unknown |
0.414 |
-0.046 |
| 88 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
0.413 |
-0.284 |
| 89 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
0.413 |
0.227 |
| 90 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.413 |
0.177 |
| 91 |
257860_at |
AT3G13062
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.412 |
0.174 |
| 92 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.408 |
-0.076 |
| 93 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.406 |
0.175 |
| 94 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.405 |
0.063 |
| 95 |
258262_at |
AT3G15770
|
unknown |
0.405 |
0.136 |
| 96 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.397 |
-0.041 |
| 97 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.395 |
-0.218 |
| 98 |
262127_at |
AT1G52550
|
unknown |
0.394 |
0.067 |
| 99 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.393 |
0.185 |
| 100 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.393 |
0.142 |
| 101 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.392 |
-0.056 |
| 102 |
264894_at |
AT1G23040
|
[hydroxyproline-rich glycoprotein family protein] |
0.390 |
0.249 |
| 103 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.387 |
0.021 |
| 104 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.386 |
0.509 |
| 105 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.384 |
0.354 |
| 106 |
245353_at |
AT4G16000
|
unknown |
0.383 |
0.076 |
| 107 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.380 |
0.108 |
| 108 |
246975_at |
AT5G24890
|
unknown |
0.377 |
0.127 |
| 109 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.376 |
0.110 |
| 110 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.374 |
0.247 |
| 111 |
258026_at |
AT3G19290
|
ABF4, ABRE binding factor 4, AREB2, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 2 |
0.374 |
-0.074 |
| 112 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.373 |
0.144 |
| 113 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
0.369 |
-0.055 |
| 114 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.366 |
-0.118 |
| 115 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.364 |
0.352 |
| 116 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.360 |
0.106 |
| 117 |
260875_at |
AT1G21410
|
SKP2A |
0.360 |
0.157 |
| 118 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
0.358 |
0.273 |
| 119 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.357 |
0.126 |
| 120 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.356 |
0.287 |
| 121 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.355 |
-0.193 |
| 122 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.354 |
0.172 |
| 123 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.349 |
0.108 |
| 124 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
0.348 |
0.165 |
| 125 |
259312_at |
AT3G05200
|
ATL6, Arabidopsis toxicos en levadura 6 |
0.347 |
0.019 |
| 126 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.591 |
-0.101 |
| 127 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.547 |
0.025 |
| 128 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
-0.508 |
-0.019 |
| 129 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.479 |
-0.058 |
| 130 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.456 |
-0.243 |
| 131 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.450 |
-0.234 |
| 132 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.440 |
-0.181 |
| 133 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
-0.436 |
-0.011 |
| 134 |
245063_at |
AT2G39795
|
[Mitochondrial glycoprotein family protein] |
-0.432 |
-0.015 |
| 135 |
253516_at |
AT4G31360
|
[selenium binding] |
-0.428 |
-0.148 |
| 136 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.425 |
-0.033 |
| 137 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
-0.412 |
-0.068 |
| 138 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
-0.399 |
0.097 |
| 139 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.398 |
-0.098 |
| 140 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.397 |
-0.034 |
| 141 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.394 |
-0.173 |
| 142 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.392 |
-0.138 |
| 143 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.383 |
-0.054 |
| 144 |
251830_at |
AT3G55010
|
ATPURM, EMB2818, EMBRYO DEFECTIVE 2818, PUR5 |
-0.377 |
-0.018 |
| 145 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.377 |
-0.127 |
| 146 |
267309_at |
AT2G19385
|
[zinc ion binding] |
-0.370 |
-0.207 |
| 147 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
-0.366 |
0.193 |
| 148 |
247575_at |
AT5G61030
|
GR-RBP3, glycine-rich RNA-binding protein 3 |
-0.365 |
-0.054 |
| 149 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
-0.363 |
0.078 |
| 150 |
266934_at |
AT2G18900
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.363 |
-0.039 |
| 151 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
-0.362 |
-0.054 |
| 152 |
258410_at |
AT3G16780
|
[Ribosomal protein L19e family protein] |
-0.361 |
-0.043 |
| 153 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
-0.357 |
-0.069 |
| 154 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
-0.353 |
-0.006 |
| 155 |
264913_at |
AT1G60770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.348 |
-0.068 |
| 156 |
247119_at |
AT5G65900
|
[DEA(D/H)-box RNA helicase family protein] |
-0.343 |
-0.041 |
| 157 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.336 |
-0.351 |
| 158 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.336 |
-0.016 |
| 159 |
246898_at |
AT5G25580
|
unknown |
-0.334 |
0.010 |
| 160 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.329 |
-0.170 |
| 161 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
-0.329 |
-0.025 |
| 162 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
-0.329 |
-0.167 |
| 163 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
-0.328 |
-0.130 |
| 164 |
253817_at |
AT4G28310
|
unknown |
-0.325 |
0.015 |
| 165 |
260984_at |
AT1G53645
|
[hydroxyproline-rich glycoprotein family protein] |
-0.321 |
-0.045 |
| 166 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
-0.321 |
-0.058 |
| 167 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.319 |
-0.108 |
| 168 |
249901_at |
AT5G22650
|
ATHD2, ARABIDOPSIS HISTONE DEACETYLASE 2, ATHD2B, HD2, HISTONE DEACETYLASE 2, HD2B, histone deacetylase 2B, HDA4, HDT02, HDT2 |
-0.316 |
0.005 |
| 169 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.316 |
-0.047 |
| 170 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
-0.316 |
-0.084 |
| 171 |
247249_at |
AT5G64670
|
[Ribosomal protein L18e/L15 superfamily protein] |
-0.315 |
-0.024 |
| 172 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
-0.314 |
0.036 |
| 173 |
255876_at |
AT2G40480
|
unknown |
-0.312 |
-0.153 |
| 174 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.311 |
-0.475 |
| 175 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.310 |
0.083 |
| 176 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.310 |
-0.279 |
| 177 |
245502_at |
AT4G15640
|
unknown |
-0.310 |
-0.102 |
| 178 |
256144_at |
AT1G48630
|
RACK1B, receptor for activated C kinase 1B, RACK1B_AT, receptor for activated C kinase 1B |
-0.309 |
0.004 |
| 179 |
246094_at |
AT5G19300
|
unknown |
-0.307 |
-0.006 |
| 180 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.307 |
-0.208 |
| 181 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.306 |
-0.064 |
| 182 |
265864_at |
AT2G01750
|
ATMAP70-3, microtubule-associated proteins 70-3, MAP70-3, microtubule-associated proteins 70-3 |
-0.306 |
-0.189 |
| 183 |
257237_at |
AT3G14890
|
[phosphoesterase] |
-0.304 |
0.013 |
| 184 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.302 |
-0.044 |
| 185 |
258155_at |
AT3G18130
|
RACK1C, receptor for activated C kinase 1C, RACK1C_AT, receptor for activated C kinase 1C |
-0.299 |
-0.057 |
| 186 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
-0.297 |
-0.141 |
| 187 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
-0.296 |
-0.161 |
| 188 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
-0.295 |
-0.052 |
| 189 |
245085_at |
AT2G23350
|
PAB4, poly(A) binding protein 4, PABP4, POLY(A) BINDING PROTEIN 4 |
-0.293 |
-0.022 |
| 190 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.293 |
0.088 |
| 191 |
247517_at |
AT5G61780
|
AtTudor2, Arabidopsis thaliana TUDOR-SN protein 2, TSN2, TUDOR-SN protein 2, Tudor2, TUDOR-SN protein 2 |
-0.293 |
-0.089 |
| 192 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
-0.292 |
0.003 |
| 193 |
250480_at |
AT5G10290
|
[leucine-rich repeat transmembrane protein kinase family protein] |
-0.288 |
0.014 |
| 194 |
253058_at |
AT4G37660
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
-0.287 |
-0.015 |
| 195 |
260683_at |
AT1G17560
|
HLL, HUELLENLOS |
-0.287 |
0.146 |
| 196 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
-0.287 |
-0.096 |
| 197 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
-0.285 |
-0.246 |
| 198 |
256253_at |
AT3G11250
|
[Ribosomal protein L10 family protein] |
-0.285 |
0.062 |
| 199 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.283 |
-0.042 |
| 200 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
-0.283 |
-0.173 |
| 201 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.282 |
-0.043 |
| 202 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.281 |
-0.063 |
| 203 |
267174_at |
AT2G37600
|
[Ribosomal protein L36e family protein] |
-0.280 |
0.050 |
| 204 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.279 |
0.024 |
| 205 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.279 |
-0.049 |
| 206 |
264675_at |
AT1G09830
|
PUR2, purine biosynthesis 2 |
-0.279 |
-0.020 |
| 207 |
253445_at |
AT4G32605
|
[Mitochondrial glycoprotein family protein] |
-0.279 |
0.088 |
| 208 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.278 |
0.023 |
| 209 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
-0.278 |
-0.010 |
| 210 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
-0.277 |
-0.141 |
| 211 |
260325_at |
AT1G63940
|
MDAR6, monodehydroascorbate reductase 6 |
-0.277 |
-0.145 |
| 212 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.276 |
0.179 |
| 213 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.276 |
-0.049 |
| 214 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.276 |
0.002 |
| 215 |
250461_at |
AT5G10010
|
HIT4, HEAT INTOLERANT 4 |
-0.276 |
-0.173 |
| 216 |
249821_at |
AT5G23690
|
[Polynucleotide adenylyltransferase family protein] |
-0.274 |
-0.051 |
| 217 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
-0.273 |
-0.088 |
| 218 |
260192_at |
AT1G67630
|
EMB2814, EMBRYO DEFECTIVE 2814, POLA2, DNA polymerase alpha 2 |
-0.272 |
-0.042 |
| 219 |
247441_at |
AT5G62710
|
[Leucine-rich repeat protein kinase family protein] |
-0.270 |
0.199 |
| 220 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
-0.269 |
-0.073 |
| 221 |
258701_at |
AT3G09720
|
AtRH57, RH57, RNA helicase 57 |
-0.269 |
0.090 |
| 222 |
263448_at |
AT2G31660
|
EMA1, enhanced miRNA activity 1, SAD2, SUPER SENSITIVE TO ABA AND DROUGHT2, URM9, UNARMED 9 |
-0.267 |
-0.063 |
| 223 |
260222_at |
AT1G74380
|
XXT5, xyloglucan xylosyltransferase 5 |
-0.266 |
-0.008 |
| 224 |
250206_at |
AT5G14040
|
MPT3, mitochondrial phosphate transporter 3, PHT3;1, phosphate transporter 3;1 |
-0.266 |
-0.019 |
| 225 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.265 |
-0.082 |
| 226 |
254831_at |
AT4G12600
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.265 |
0.011 |
| 227 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
-0.265 |
-0.039 |
| 228 |
258104_at |
AT3G23620
|
[Ribosomal RNA processing Brix domain protein] |
-0.264 |
-0.074 |
| 229 |
258220_at |
AT3G17830
|
DJA4, DNA J protein A4 |
-0.263 |
0.055 |
| 230 |
254684_at |
AT4G13850
|
ATGRP2, GLYCINE-RICH RNA-BINDING PROTEIN 2, GR-RBP2, glycine-rich RNA-binding protein 2, GRP2, glycine rich protein 2 |
-0.262 |
-0.023 |
| 231 |
250309_at |
AT5G12220
|
[las1-like family protein] |
-0.262 |
-0.071 |
| 232 |
258482_at |
AT3G02530
|
[TCP-1/cpn60 chaperonin family protein] |
-0.262 |
0.039 |
| 233 |
250484_at |
AT5G10240
|
ASN3, asparagine synthetase 3 |
-0.261 |
0.017 |
| 234 |
251574_at |
AT3G58100
|
PDCB5, plasmodesmata callose-binding protein 5 |
-0.261 |
0.004 |
| 235 |
248341_at |
AT5G52220
|
unknown |
-0.260 |
0.077 |
| 236 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.260 |
0.048 |
| 237 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
-0.256 |
-0.154 |
| 238 |
254991_at |
AT4G10620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.255 |
-0.024 |
| 239 |
258816_at |
AT3G03960
|
[TCP-1/cpn60 chaperonin family protein] |
-0.253 |
0.031 |
| 240 |
258692_at |
AT3G08640
|
RER3, RETICULATA-RELATED 3 |
-0.253 |
-0.012 |
| 241 |
265667_at |
AT2G27470
|
NF-YB11, nuclear factor Y, subunit B11 |
-0.253 |
-0.158 |
| 242 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.253 |
0.011 |
| 243 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.253 |
-0.209 |
| 244 |
250779_at |
AT5G05470
|
ATEIF2-A2, EIF2 ALPHA, eukaryotic translation initiation factor 2 alpha subunit, EIF2-A2 |
-0.253 |
-0.132 |
| 245 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.252 |
-0.177 |
| 246 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.252 |
-0.036 |
| 247 |
267586_at |
AT2G41950
|
unknown |
-0.251 |
0.010 |
| 248 |
265389_at |
AT2G20690
|
[lumazine-binding family protein] |
-0.251 |
-0.181 |
| 249 |
250084_at |
AT5G17240
|
SDG40, SET domain group 40 |
-0.248 |
0.100 |