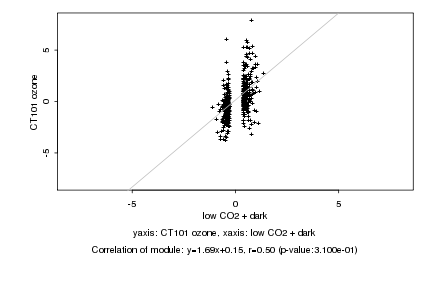
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
low CO2 + dark |
Log2 signal ratio
CT101 ozone |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.313 |
2.743 |
| 2 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.125 |
1.047 |
| 3 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
1.090 |
-2.046 |
| 4 |
252882_at |
AT4G39675
|
unknown |
1.021 |
1.990 |
| 5 |
264580_at |
AT1G05340
|
unknown |
1.020 |
3.696 |
| 6 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
1.018 |
1.406 |
| 7 |
264238_at |
AT1G54740
|
unknown |
1.003 |
-0.916 |
| 8 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.975 |
2.413 |
| 9 |
265276_at |
AT2G28400
|
unknown |
0.969 |
4.468 |
| 10 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.962 |
0.950 |
| 11 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.958 |
3.669 |
| 12 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.948 |
3.391 |
| 13 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.889 |
-2.039 |
| 14 |
249454_at |
AT5G39520
|
unknown |
0.886 |
-0.794 |
| 15 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.863 |
0.790 |
| 16 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.835 |
3.225 |
| 17 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.822 |
1.233 |
| 18 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.812 |
1.141 |
| 19 |
263881_at |
AT2G21820
|
unknown |
0.810 |
-0.147 |
| 20 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.806 |
1.095 |
| 21 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.806 |
1.169 |
| 22 |
264774_at |
AT1G22890
|
unknown |
0.802 |
4.693 |
| 23 |
247835_at |
AT5G57910
|
unknown |
0.800 |
3.230 |
| 24 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.795 |
1.907 |
| 25 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.788 |
5.384 |
| 26 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.780 |
0.740 |
| 27 |
256799_at |
AT3G18560
|
unknown |
0.775 |
2.940 |
| 28 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.756 |
0.294 |
| 29 |
265387_at |
AT2G20670
|
unknown |
0.754 |
-2.177 |
| 30 |
253827_at |
AT4G28085
|
unknown |
0.751 |
7.976 |
| 31 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.735 |
1.914 |
| 32 |
254193_at |
AT4G23870
|
unknown |
0.731 |
-3.196 |
| 33 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.709 |
4.107 |
| 34 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.705 |
-1.767 |
| 35 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.700 |
2.482 |
| 36 |
259982_at |
AT1G76410
|
ATL8 |
0.697 |
2.636 |
| 37 |
266259_at |
AT2G27830
|
unknown |
0.691 |
0.593 |
| 38 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.686 |
0.057 |
| 39 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.680 |
0.225 |
| 40 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.675 |
-2.579 |
| 41 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.663 |
1.443 |
| 42 |
258939_at |
AT3G10020
|
unknown |
0.660 |
0.187 |
| 43 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.656 |
0.312 |
| 44 |
260005_at |
AT1G67920
|
unknown |
0.655 |
5.166 |
| 45 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.654 |
4.724 |
| 46 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.652 |
0.961 |
| 47 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
0.649 |
2.267 |
| 48 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.649 |
0.564 |
| 49 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.648 |
1.142 |
| 50 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.646 |
1.742 |
| 51 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.634 |
2.222 |
| 52 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.631 |
-1.367 |
| 53 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.631 |
0.614 |
| 54 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.626 |
0.661 |
| 55 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.624 |
0.737 |
| 56 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.620 |
2.674 |
| 57 |
259015_at |
AT3G07350
|
unknown |
0.616 |
-1.801 |
| 58 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.610 |
-0.308 |
| 59 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.605 |
-0.346 |
| 60 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.601 |
-0.963 |
| 61 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.601 |
-0.079 |
| 62 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.597 |
-1.017 |
| 63 |
259856_at |
AT1G68440
|
unknown |
0.585 |
-0.068 |
| 64 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.579 |
0.881 |
| 65 |
262801_at |
AT1G21010
|
unknown |
0.565 |
2.136 |
| 66 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.558 |
0.937 |
| 67 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.555 |
3.444 |
| 68 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.555 |
5.282 |
| 69 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.554 |
-0.467 |
| 70 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.554 |
0.836 |
| 71 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.552 |
-0.982 |
| 72 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
0.550 |
0.338 |
| 73 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.547 |
-0.664 |
| 74 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.544 |
1.517 |
| 75 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.541 |
1.346 |
| 76 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.537 |
4.285 |
| 77 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.536 |
5.742 |
| 78 |
264447_at |
AT1G27300
|
unknown |
0.534 |
0.516 |
| 79 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.533 |
1.841 |
| 80 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.531 |
5.943 |
| 81 |
261567_at |
AT1G33055
|
unknown |
0.525 |
2.185 |
| 82 |
246951_at |
AT5G04880
|
[expressed protein] |
0.522 |
-0.712 |
| 83 |
258262_at |
AT3G15770
|
unknown |
0.522 |
1.549 |
| 84 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.520 |
1.728 |
| 85 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.519 |
2.276 |
| 86 |
258209_at |
AT3G14060
|
unknown |
0.518 |
0.629 |
| 87 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.517 |
-0.501 |
| 88 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.515 |
-0.819 |
| 89 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.513 |
-0.231 |
| 90 |
246506_at |
AT5G16110
|
unknown |
0.512 |
1.091 |
| 91 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.509 |
4.413 |
| 92 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.508 |
0.883 |
| 93 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.507 |
4.642 |
| 94 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.507 |
-0.135 |
| 95 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.506 |
2.452 |
| 96 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.506 |
0.349 |
| 97 |
262803_at |
AT1G21000
|
[PLATZ transcription factor family protein] |
0.506 |
1.108 |
| 98 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.503 |
4.387 |
| 99 |
256442_at |
AT3G10930
|
unknown |
0.503 |
3.659 |
| 100 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.497 |
5.333 |
| 101 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.496 |
0.225 |
| 102 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.495 |
1.923 |
| 103 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
0.489 |
-0.502 |
| 104 |
265470_at |
AT2G37150
|
[RING/U-box superfamily protein] |
0.481 |
2.530 |
| 105 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.479 |
1.701 |
| 106 |
261205_at |
AT1G12790
|
unknown |
0.477 |
3.584 |
| 107 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.477 |
0.204 |
| 108 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
0.475 |
1.703 |
| 109 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.475 |
-0.698 |
| 110 |
246975_at |
AT5G24890
|
unknown |
0.467 |
-0.242 |
| 111 |
261615_at |
AT1G33050
|
unknown |
0.466 |
1.361 |
| 112 |
254787_at |
AT4G12690
|
unknown |
0.466 |
0.008 |
| 113 |
258275_at |
AT3G15760
|
unknown |
0.464 |
1.742 |
| 114 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
0.463 |
-0.428 |
| 115 |
258647_at |
AT3G07870
|
[F-box and associated interaction domains-containing protein] |
0.463 |
1.426 |
| 116 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
0.458 |
-0.054 |
| 117 |
266195_at |
AT2G39100
|
[RING/U-box superfamily protein] |
0.458 |
0.617 |
| 118 |
255285_at |
AT4G04630
|
unknown |
0.455 |
0.481 |
| 119 |
253147_at |
AT4G35600
|
CST, CAST AWAY, CX32, CONNEXIN 32, Kin4, kinase 4 |
0.455 |
3.450 |
| 120 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.454 |
1.195 |
| 121 |
264423_at |
AT1G61690
|
[phosphoinositide binding] |
0.454 |
1.729 |
| 122 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.453 |
0.501 |
| 123 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.448 |
0.225 |
| 124 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.448 |
1.354 |
| 125 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.447 |
3.606 |
| 126 |
249824_at |
AT5G23380
|
unknown |
0.447 |
0.449 |
| 127 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.445 |
0.783 |
| 128 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
0.431 |
0.941 |
| 129 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.431 |
2.468 |
| 130 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
0.430 |
-1.214 |
| 131 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.429 |
1.923 |
| 132 |
264379_at |
AT2G25200
|
unknown |
0.427 |
-1.254 |
| 133 |
252323_at |
AT3G48530
|
KING1, SNF1-related protein kinase regulatory subunit gamma 1 |
0.427 |
-0.556 |
| 134 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.427 |
3.530 |
| 135 |
264836_at |
AT1G03610
|
unknown |
0.426 |
-0.163 |
| 136 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.425 |
1.863 |
| 137 |
259996_at |
AT1G67910
|
unknown |
0.423 |
-0.293 |
| 138 |
252057_at |
AT3G52480
|
unknown |
0.422 |
1.738 |
| 139 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.420 |
0.133 |
| 140 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
0.418 |
0.571 |
| 141 |
262049_at |
AT1G80180
|
unknown |
0.416 |
0.555 |
| 142 |
249140_at |
AT5G43190
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.416 |
3.283 |
| 143 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.415 |
-2.367 |
| 144 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.414 |
0.778 |
| 145 |
260310_at |
AT1G70590
|
[F-box family protein] |
0.413 |
1.105 |
| 146 |
261253_at |
AT1G05840
|
[Eukaryotic aspartyl protease family protein] |
0.412 |
0.782 |
| 147 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.411 |
-0.522 |
| 148 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.410 |
-0.439 |
| 149 |
260875_at |
AT1G21410
|
SKP2A |
0.410 |
-1.222 |
| 150 |
245781_at |
AT1G45976
|
SBP1, S-ribonuclease binding protein 1 |
0.408 |
0.604 |
| 151 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.408 |
-0.692 |
| 152 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.405 |
0.440 |
| 153 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.401 |
0.343 |
| 154 |
248901_at |
AT5G46410
|
SSP4, SCP1-like small phosphatase 4 |
0.400 |
1.336 |
| 155 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.396 |
1.325 |
| 156 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.396 |
0.510 |
| 157 |
245776_at |
AT1G30260
|
unknown |
0.395 |
-1.398 |
| 158 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.394 |
-0.187 |
| 159 |
259769_at |
AT1G29400
|
AML5, MEI2-like protein 5, ML5, MEI2-like protein 5 |
0.393 |
0.180 |
| 160 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.390 |
-0.337 |
| 161 |
253808_at |
AT4G28300
|
unknown |
0.390 |
1.023 |
| 162 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.390 |
2.039 |
| 163 |
262607_at |
AT1G13990
|
unknown |
0.388 |
2.210 |
| 164 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
0.387 |
-0.708 |
| 165 |
260544_at |
AT2G43540
|
unknown |
0.386 |
-0.282 |
| 166 |
264772_at |
AT1G22930
|
[T-complex protein 11] |
0.386 |
0.995 |
| 167 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
0.385 |
-0.594 |
| 168 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.385 |
0.693 |
| 169 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.383 |
0.987 |
| 170 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.383 |
2.285 |
| 171 |
258565_at |
AT3G04350
|
unknown |
0.382 |
0.382 |
| 172 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.381 |
1.233 |
| 173 |
247585_at |
AT5G60680
|
unknown |
0.381 |
-1.213 |
| 174 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.381 |
0.651 |
| 175 |
262992_at |
AT1G54210
|
APG12, AUTOPHAGY 12, ATATG12, ATG12A, AUTOPHAGY 12 A |
0.380 |
0.737 |
| 176 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.380 |
5.336 |
| 177 |
250327_at |
AT5G12050
|
unknown |
0.380 |
-1.784 |
| 178 |
253716_at |
AT4G29420
|
[F-box/RNI-like superfamily protein] |
0.378 |
0.468 |
| 179 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.378 |
0.057 |
| 180 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.378 |
0.130 |
| 181 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
0.377 |
0.122 |
| 182 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.377 |
0.796 |
| 183 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.377 |
-1.757 |
| 184 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
0.376 |
-0.778 |
| 185 |
261250_at |
AT1G05890
|
ARI5, ARIADNE 5, ATARI5, ARABIDOPSIS ARIADNE 5 |
0.375 |
1.015 |
| 186 |
250028_at |
AT5G18130
|
unknown |
0.375 |
3.104 |
| 187 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.375 |
-1.046 |
| 188 |
264304_at |
AT1G78895
|
[Reticulon family protein] |
0.373 |
2.553 |
| 189 |
253958_at |
AT4G26400
|
[RING/U-box superfamily protein] |
0.371 |
2.318 |
| 190 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
0.370 |
0.027 |
| 191 |
252424_at |
AT3G47610
|
[transcription regulators] |
0.368 |
-0.047 |
| 192 |
264508_at |
AT1G09570
|
FHY2, FAR RED ELONGATED HYPOCOTYL 2, FRE1, FAR RED ELONGATED 1, HY8, ELONGATED HYPOCOTYL 8, PHYA, phytochrome A |
0.368 |
-0.299 |
| 193 |
245353_at |
AT4G16000
|
unknown |
0.367 |
1.358 |
| 194 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
0.365 |
2.217 |
| 195 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
0.365 |
0.782 |
| 196 |
248879_at |
AT5G46180
|
DELTA-OAT, ornithine-delta-aminotransferase |
0.364 |
-0.067 |
| 197 |
264512_at |
AT1G09575
|
unknown |
0.363 |
3.537 |
| 198 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.363 |
1.481 |
| 199 |
267375_at |
AT2G26300
|
ATGPA1, ARABIDOPSIS THALIANA G PROTEIN ALPHA SUBUNIT 1, GP ALPHA 1, G protein alpha subunit 1, GPA1, G PROTEIN ALPHA SUBUNIT 1 |
0.362 |
1.141 |
| 200 |
249345_at |
AT5G40740
|
AUG6, augmin subunit 6 |
0.362 |
0.103 |
| 201 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.362 |
1.550 |
| 202 |
260486_at |
AT1G51550
|
[Kelch repeat-containing F-box family protein] |
0.362 |
-0.434 |
| 203 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.361 |
0.359 |
| 204 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.361 |
1.753 |
| 205 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.360 |
-0.099 |
| 206 |
258026_at |
AT3G19290
|
ABF4, ABRE binding factor 4, AREB2, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 2 |
0.358 |
0.262 |
| 207 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.358 |
-0.460 |
| 208 |
266811_at |
AT2G44850
|
unknown |
0.357 |
-0.236 |
| 209 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.356 |
1.570 |
| 210 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
0.355 |
-0.859 |
| 211 |
263839_at |
AT2G36900
|
ATMEMB11, MEMB11, membrin 11 |
0.353 |
1.483 |
| 212 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
0.352 |
-2.070 |
| 213 |
260094_at |
AT1G73250
|
ATFX, ACTIVATING TRANSCRIPTION FACTOR 5, GER1, GDP-4-keto-6-deoxymannose-3,5-epimerase-4-reductase 1 |
0.352 |
1.524 |
| 214 |
250830_at |
AT5G04910
|
unknown |
0.352 |
0.160 |
| 215 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
0.351 |
3.831 |
| 216 |
258953_at |
AT3G01430
|
unknown |
0.350 |
0.745 |
| 217 |
249650_at |
AT5G37055
|
ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING |
0.350 |
0.418 |
| 218 |
250000_at |
AT5G18650
|
MIEL1, MYB30-Interacting E3 Ligase 1 |
0.349 |
-0.023 |
| 219 |
262166_at |
AT1G74840
|
[Homeodomain-like superfamily protein] |
0.349 |
-1.207 |
| 220 |
252661_at |
AT3G44450
|
unknown |
-1.141 |
-0.548 |
| 221 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.964 |
-1.668 |
| 222 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.905 |
-2.966 |
| 223 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.836 |
-0.242 |
| 224 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.812 |
-0.933 |
| 225 |
251058_at |
AT5G01790
|
unknown |
-0.765 |
-3.386 |
| 226 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.733 |
-0.761 |
| 227 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.733 |
-3.618 |
| 228 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.705 |
-2.881 |
| 229 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.671 |
-0.448 |
| 230 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.647 |
-1.864 |
| 231 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.642 |
0.142 |
| 232 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.632 |
-1.697 |
| 233 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.632 |
-1.949 |
| 234 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.628 |
-0.496 |
| 235 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.621 |
-2.492 |
| 236 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
-0.614 |
0.018 |
| 237 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.601 |
-0.285 |
| 238 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.600 |
-3.636 |
| 239 |
252010_at |
AT3G52740
|
unknown |
-0.599 |
-2.779 |
| 240 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.597 |
-1.590 |
| 241 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.594 |
-1.448 |
| 242 |
256617_at |
AT3G22240
|
unknown |
-0.591 |
2.067 |
| 243 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.586 |
1.612 |
| 244 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.583 |
-1.827 |
| 245 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.583 |
-0.325 |
| 246 |
266384_at |
AT2G14660
|
unknown |
-0.580 |
-1.792 |
| 247 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.571 |
1.356 |
| 248 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
-0.569 |
-0.512 |
| 249 |
248139_at |
AT5G54970
|
unknown |
-0.563 |
0.711 |
| 250 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.561 |
-0.910 |
| 251 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.559 |
-2.056 |
| 252 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.556 |
-0.751 |
| 253 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.554 |
-0.709 |
| 254 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.553 |
-0.567 |
| 255 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.550 |
-1.542 |
| 256 |
251476_at |
AT3G59670
|
unknown |
-0.546 |
-0.533 |
| 257 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.536 |
-0.777 |
| 258 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.531 |
-1.953 |
| 259 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.531 |
-0.573 |
| 260 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.530 |
-0.501 |
| 261 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.530 |
-2.332 |
| 262 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
-0.529 |
-0.611 |
| 263 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.527 |
0.223 |
| 264 |
253777_at |
AT4G28450
|
[nucleotide binding] |
-0.526 |
0.242 |
| 265 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.525 |
-0.814 |
| 266 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.524 |
-0.555 |
| 267 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.516 |
-1.283 |
| 268 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.515 |
-1.220 |
| 269 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.514 |
-1.522 |
| 270 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
-0.512 |
-0.114 |
| 271 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.507 |
0.172 |
| 272 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.503 |
0.419 |
| 273 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.503 |
-3.737 |
| 274 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.497 |
-0.653 |
| 275 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.493 |
-1.100 |
| 276 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.492 |
-0.607 |
| 277 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.489 |
0.281 |
| 278 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.486 |
-0.246 |
| 279 |
245063_at |
AT2G39795
|
[Mitochondrial glycoprotein family protein] |
-0.486 |
-0.312 |
| 280 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.486 |
-0.727 |
| 281 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.485 |
-3.398 |
| 282 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.484 |
0.199 |
| 283 |
266338_at |
AT2G32400
|
ATGLR3.7, GLR3.7, GLUTAMATE RECEPTOR 3.7, GLR5, glutamate receptor 5 |
-0.484 |
-0.009 |
| 284 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
-0.482 |
-0.827 |
| 285 |
251753_at |
AT3G55760
|
unknown |
-0.482 |
-1.784 |
| 286 |
261772_at |
AT1G76240
|
unknown |
-0.480 |
-1.787 |
| 287 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.478 |
-1.106 |
| 288 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.476 |
0.225 |
| 289 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.476 |
0.380 |
| 290 |
267044_at |
AT2G34357
|
[ARM repeat superfamily protein] |
-0.475 |
-0.298 |
| 291 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
-0.475 |
-0.043 |
| 292 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
-0.474 |
-0.116 |
| 293 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.472 |
-3.460 |
| 294 |
262094_at |
AT1G56110
|
NOP56, homolog of nucleolar protein NOP56 |
-0.471 |
-0.287 |
| 295 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.471 |
3.809 |
| 296 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.468 |
-2.146 |
| 297 |
246315_at |
AT3G56870
|
unknown |
-0.467 |
0.531 |
| 298 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
-0.462 |
-1.900 |
| 299 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
-0.460 |
-0.468 |
| 300 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.455 |
-0.562 |
| 301 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
-0.454 |
0.500 |
| 302 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
-0.454 |
-2.318 |
| 303 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
-0.454 |
0.145 |
| 304 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
-0.451 |
-0.362 |
| 305 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.448 |
-0.550 |
| 306 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.447 |
6.075 |
| 307 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
-0.447 |
-0.759 |
| 308 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.444 |
-1.610 |
| 309 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.442 |
-1.178 |
| 310 |
246266_at |
AT1G31817
|
NFD3, NUCLEAR FUSION DEFECTIVE 3 |
-0.440 |
-0.102 |
| 311 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
-0.439 |
-0.375 |
| 312 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.437 |
-0.997 |
| 313 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.436 |
1.368 |
| 314 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.434 |
-0.625 |
| 315 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.434 |
1.696 |
| 316 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
-0.433 |
-0.887 |
| 317 |
267004_at |
AT2G34260
|
WDR55, human WDR55 (WD40 repeat) homolog |
-0.433 |
-0.322 |
| 318 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.431 |
1.599 |
| 319 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.429 |
-2.862 |
| 320 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
-0.427 |
0.915 |
| 321 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.427 |
-0.683 |
| 322 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.423 |
-0.248 |
| 323 |
247420_at |
AT5G63100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.422 |
-1.445 |
| 324 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
-0.421 |
-0.324 |
| 325 |
265340_at |
AT2G18330
|
[AAA-type ATPase family protein] |
-0.420 |
0.838 |
| 326 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
-0.420 |
-0.311 |
| 327 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.418 |
-0.839 |
| 328 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
-0.418 |
0.279 |
| 329 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.418 |
-1.921 |
| 330 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.417 |
-0.552 |
| 331 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.417 |
-0.547 |
| 332 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
-0.416 |
0.048 |
| 333 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.413 |
-1.478 |
| 334 |
246767_at |
AT5G27395
|
[Mitochondrial inner membrane translocase complex, subunit Tim44-related protein] |
-0.413 |
-0.155 |
| 335 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.413 |
-0.661 |
| 336 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
-0.412 |
-0.638 |
| 337 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.411 |
-1.241 |
| 338 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
-0.411 |
-0.490 |
| 339 |
251830_at |
AT3G55010
|
ATPURM, EMB2818, EMBRYO DEFECTIVE 2818, PUR5 |
-0.410 |
-0.694 |
| 340 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.409 |
-0.595 |
| 341 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.408 |
-1.716 |
| 342 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
-0.406 |
0.520 |
| 343 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.405 |
0.354 |
| 344 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.405 |
-1.617 |
| 345 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.404 |
-0.610 |
| 346 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.398 |
-0.872 |
| 347 |
254455_at |
AT4G21140
|
unknown |
-0.394 |
0.409 |
| 348 |
261921_at |
AT1G65900
|
unknown |
-0.393 |
-1.467 |
| 349 |
250394_at |
AT5G10910
|
[mraW methylase family protein] |
-0.393 |
-0.970 |
| 350 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.391 |
-1.844 |
| 351 |
262738_at |
AT1G28530
|
unknown |
-0.389 |
-0.937 |
| 352 |
262933_at |
AT1G65840
|
ATPAO4, polyamine oxidase 4, PAO4, polyamine oxidase 4 |
-0.388 |
1.186 |
| 353 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.388 |
-3.078 |
| 354 |
245750_at |
AT1G51060
|
HTA10, histone H2A 10 |
-0.388 |
-0.144 |
| 355 |
251760_at |
AT3G55605
|
[Mitochondrial glycoprotein family protein] |
-0.388 |
-0.386 |
| 356 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.387 |
-0.358 |
| 357 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
-0.386 |
-0.279 |
| 358 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
-0.385 |
-0.020 |
| 359 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.385 |
2.992 |
| 360 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.384 |
-1.017 |
| 361 |
265704_at |
AT2G03420
|
unknown |
-0.384 |
0.225 |
| 362 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
-0.382 |
-0.089 |
| 363 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.381 |
-0.647 |
| 364 |
267586_at |
AT2G41950
|
unknown |
-0.381 |
-1.556 |
| 365 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.380 |
-0.838 |
| 366 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
-0.379 |
-0.170 |
| 367 |
247575_at |
AT5G61030
|
GR-RBP3, glycine-rich RNA-binding protein 3 |
-0.379 |
-0.574 |
| 368 |
261810_at |
AT1G08130
|
ATLIG1, DNA ligase 1, LIG1, DNA ligase 1 |
-0.379 |
-0.272 |
| 369 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.378 |
1.437 |
| 370 |
255465_at |
AT4G02990
|
BSM, BELAYA SMERT, RUG2, RUGOSA 2 |
-0.378 |
-0.941 |
| 371 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.378 |
0.099 |
| 372 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.378 |
-2.264 |
| 373 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
-0.378 |
-0.605 |
| 374 |
266478_at |
AT2G31170
|
FIONA, FIONA, SYCO ARATH, cysteinyl t-RNA synthetase |
-0.377 |
-0.929 |
| 375 |
261411_at |
AT1G07790
|
HTB1 |
-0.376 |
-0.600 |
| 376 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.376 |
-0.512 |
| 377 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.375 |
-2.912 |
| 378 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.373 |
-0.144 |
| 379 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.373 |
-0.629 |
| 380 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
-0.370 |
-0.594 |
| 381 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.369 |
-2.015 |
| 382 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.369 |
-0.545 |
| 383 |
266567_at |
AT2G24050
|
eIFiso4G2, eukaryotic translation Initiation Factor isoform 4G2 |
-0.369 |
-0.371 |
| 384 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
-0.369 |
-0.250 |
| 385 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.369 |
-2.275 |
| 386 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
-0.369 |
-0.673 |
| 387 |
251415_at |
AT3G60380
|
unknown |
-0.368 |
-1.391 |
| 388 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.368 |
-2.360 |
| 389 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.367 |
1.101 |
| 390 |
265474_at |
AT2G15690
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.367 |
-0.302 |
| 391 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.365 |
-1.630 |
| 392 |
248889_at |
AT5G46230
|
unknown |
-0.363 |
2.685 |
| 393 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.361 |
-1.479 |
| 394 |
265309_at |
AT2G20290
|
ATXIG, MYOSIN XI G, XIG, myosin-like protein XIG |
-0.359 |
-1.368 |
| 395 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.355 |
-1.207 |
| 396 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
-0.354 |
-0.501 |
| 397 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
-0.352 |
-1.600 |
| 398 |
264386_at |
AT1G12000
|
[Phosphofructokinase family protein] |
-0.352 |
0.166 |
| 399 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
-0.351 |
-0.172 |
| 400 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.350 |
-1.084 |
| 401 |
261661_at |
AT1G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.349 |
-1.764 |
| 402 |
246094_at |
AT5G19300
|
unknown |
-0.349 |
0.542 |
| 403 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
-0.349 |
2.273 |
| 404 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.349 |
-0.072 |
| 405 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.347 |
-0.758 |
| 406 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.347 |
-1.568 |
| 407 |
263955_at |
AT2G36010
|
ATE2FA, E2F3, E2F transcription factor 3 |
-0.346 |
0.325 |
| 408 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
-0.345 |
-0.520 |
| 409 |
258549_at |
AT3G06930
|
ATPRMT4B, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 4B, PRMT4B, protein arginine methyltransferase 4B |
-0.345 |
-0.517 |
| 410 |
259298_at |
AT3G05370
|
AtRLP31, receptor like protein 31, RLP31, receptor like protein 31 |
-0.345 |
1.847 |
| 411 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.344 |
-0.934 |
| 412 |
245094_at |
AT2G40840
|
DPE2, disproportionating enzyme 2 |
-0.343 |
-2.042 |
| 413 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
-0.343 |
2.158 |
| 414 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
-0.342 |
-1.605 |
| 415 |
247494_at |
AT5G61790
|
ATCNX1, CNX1, calnexin 1 |
-0.342 |
1.255 |
| 416 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.341 |
-0.289 |
| 417 |
255455_at |
AT4G02930
|
[GTP binding Elongation factor Tu family protein] |
-0.340 |
0.953 |
| 418 |
264002_at |
AT2G22360
|
DJA6, DNA J protein A6 |
-0.340 |
-0.784 |
| 419 |
263766_at |
AT2G21440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.340 |
-0.848 |
| 420 |
249528_at |
AT5G38720
|
unknown |
-0.340 |
0.292 |
| 421 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.339 |
-0.815 |
| 422 |
256862_at |
AT3G23940
|
[dehydratase family] |
-0.336 |
-0.470 |
| 423 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.336 |
-1.635 |
| 424 |
258996_at |
AT3G01800
|
[Ribosome recycling factor] |
-0.336 |
0.586 |
| 425 |
247425_at |
AT5G62550
|
unknown |
-0.336 |
-0.388 |
| 426 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
-0.336 |
0.998 |
| 427 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.335 |
-2.419 |
| 428 |
250009_at |
AT5G18440
|
AtNUFIP, NUFIP, nuclear FMRP-interacting protein |
-0.335 |
-0.934 |
| 429 |
266587_at |
AT2G14880
|
[SWIB/MDM2 domain superfamily protein] |
-0.334 |
-0.847 |
| 430 |
253695_at |
AT4G29510
|
ATPRMT11, ARABIDOPSIS THALIANA ARGININE METHYLTRANSFERASE 11, ATPRMT1B, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1B, PRMT11, arginine methyltransferase 11, PRMT1B, PROTEIN ARGININE METHYLTRANSFERASE 1B |
-0.334 |
-0.424 |
| 431 |
266934_at |
AT2G18900
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.334 |
0.453 |
| 432 |
261439_at |
AT1G28395
|
unknown |
-0.334 |
-0.406 |
| 433 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.333 |
0.324 |
| 434 |
266093_at |
AT2G37990
|
[ribosome biogenesis regulatory protein (RRS1) family protein] |
-0.333 |
-0.241 |
| 435 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.333 |
-1.210 |
| 436 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.332 |
-1.771 |
| 437 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
-0.332 |
0.788 |