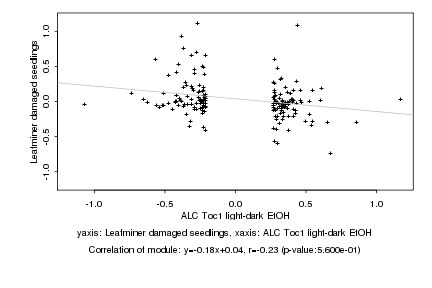
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ALC Toc1 light-dark EtOH |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
248052_at |
AT5G55800
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.167 |
0.032 |
| 2 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
0.854 |
-0.298 |
| 3 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.673 |
-0.738 |
| 4 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.651 |
-0.292 |
| 5 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.609 |
0.186 |
| 6 |
260688_at |
AT1G17665
|
unknown |
0.602 |
0.022 |
| 7 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.543 |
-0.279 |
| 8 |
267459_at |
AT2G33850
|
unknown |
0.542 |
0.171 |
| 9 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.536 |
-0.337 |
| 10 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.521 |
-0.175 |
| 11 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
0.521 |
0.001 |
| 12 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.492 |
-0.284 |
| 13 |
265611_at |
AT2G25510
|
unknown |
0.462 |
-0.009 |
| 14 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.461 |
0.171 |
| 15 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
0.459 |
0.020 |
| 16 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.438 |
1.085 |
| 17 |
251167_at |
AT3G63360
|
[defensin-related] |
0.431 |
-0.028 |
| 18 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.427 |
0.285 |
| 19 |
266592_at |
AT2G46210
|
AtSLD2, SLD2, sphingoid LCB desaturase 2 |
0.422 |
-0.166 |
| 20 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
0.421 |
-0.115 |
| 21 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.411 |
0.015 |
| 22 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.410 |
0.157 |
| 23 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
0.407 |
-0.213 |
| 24 |
249366_at |
AT5G40610
|
GPDHp, Glycerol-3-phosphate dehydrogenase plastidic |
0.407 |
-0.101 |
| 25 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.404 |
0.027 |
| 26 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.400 |
-0.003 |
| 27 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.400 |
0.035 |
| 28 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.391 |
0.034 |
| 29 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
0.387 |
0.122 |
| 30 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.379 |
0.012 |
| 31 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
0.372 |
-0.404 |
| 32 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.371 |
-0.199 |
| 33 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.371 |
-0.040 |
| 34 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.368 |
-0.013 |
| 35 |
245880_at |
AT5G09430
|
[alpha/beta-Hydrolases superfamily protein] |
0.362 |
0.132 |
| 36 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.362 |
-0.098 |
| 37 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.352 |
-0.007 |
| 38 |
259766_at |
AT1G64360
|
unknown |
0.349 |
0.209 |
| 39 |
248693_at |
AT5G48330
|
RUG2, RCC1/UVR8/GEF-like 2 |
0.348 |
-0.070 |
| 40 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.348 |
-0.032 |
| 41 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.345 |
-0.062 |
| 42 |
260937_at |
AT1G45160
|
[Protein kinase superfamily protein] |
0.342 |
-0.071 |
| 43 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.338 |
-0.143 |
| 44 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
0.334 |
-0.213 |
| 45 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
0.333 |
-0.037 |
| 46 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.331 |
-0.058 |
| 47 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
0.331 |
0.004 |
| 48 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.329 |
-0.015 |
| 49 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
0.328 |
0.024 |
| 50 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.327 |
-0.043 |
| 51 |
256125_at |
AT1G18250
|
ATLP-1 |
0.327 |
-0.255 |
| 52 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.321 |
-0.096 |
| 53 |
251238_at |
AT3G62430
|
[Protein with RNI-like/FBD-like domains] |
0.320 |
-0.116 |
| 54 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.320 |
0.333 |
| 55 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.319 |
0.318 |
| 56 |
258548_at |
AT3G06910
|
AtULP1a, ELS1, ESD4-LIKE SUMO PROTEASE 1, ULP1A, UB-like protease 1A |
0.316 |
-0.160 |
| 57 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.316 |
0.104 |
| 58 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.311 |
-0.311 |
| 59 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
0.310 |
0.052 |
| 60 |
266735_at |
AT2G46930
|
[Pectinacetylesterase family protein] |
0.306 |
-0.065 |
| 61 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.303 |
-0.014 |
| 62 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.298 |
0.474 |
| 63 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
0.296 |
-0.012 |
| 64 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.295 |
-0.088 |
| 65 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
0.292 |
-0.208 |
| 66 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.291 |
-0.588 |
| 67 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.289 |
-0.394 |
| 68 |
263284_at |
AT2G36100
|
CASP1, Casparian strip membrane domain protein 1 |
0.288 |
-0.007 |
| 69 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.288 |
-0.024 |
| 70 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
0.286 |
-0.242 |
| 71 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
0.283 |
-0.211 |
| 72 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.283 |
0.006 |
| 73 |
254205_at |
AT4G24170
|
[ATP binding microtubule motor family protein] |
0.282 |
-0.003 |
| 74 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.278 |
0.097 |
| 75 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
0.278 |
-0.127 |
| 76 |
266221_at |
AT2G28760
|
UXS6, UDP-XYL synthase 6 |
0.276 |
-0.106 |
| 77 |
260012_at |
AT1G67865
|
unknown |
0.276 |
0.169 |
| 78 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.274 |
0.267 |
| 79 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.271 |
0.609 |
| 80 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.270 |
-0.559 |
| 81 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.270 |
0.137 |
| 82 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
0.270 |
0.074 |
| 83 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
0.270 |
0.031 |
| 84 |
247210_at |
AT5G65020
|
ANNAT2, annexin 2, AtANN2 |
0.268 |
0.284 |
| 85 |
254544_at |
AT4G19820
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
0.267 |
-0.375 |
| 86 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
0.266 |
-0.071 |
| 87 |
253008_at |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
0.265 |
-0.051 |
| 88 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.265 |
0.061 |
| 89 |
259966_at |
AT1G76500
|
AHL29, AT-hook motif nuclear-localized protein 29, SOB3, SUPPRESSOR OF PHYB-4#3 |
0.264 |
-0.117 |
| 90 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
0.264 |
-0.014 |
| 91 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.264 |
-0.054 |
| 92 |
248810_at |
AT5G47280
|
ADR1-L3, ADR1-like 3 |
-1.071 |
-0.036 |
| 93 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.743 |
0.123 |
| 94 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.652 |
0.039 |
| 95 |
250135_at |
AT5G15360
|
unknown |
-0.627 |
-0.011 |
| 96 |
249454_at |
AT5G39520
|
unknown |
-0.567 |
0.603 |
| 97 |
253689_at |
AT4G29770
|
unknown |
-0.563 |
-0.047 |
| 98 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.540 |
-0.079 |
| 99 |
257417_at |
AT1G10110
|
[F-box family protein] |
-0.519 |
-0.054 |
| 100 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.511 |
-0.053 |
| 101 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
-0.510 |
0.125 |
| 102 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.480 |
0.384 |
| 103 |
255414_at |
AT4G03156
|
[small GTPase-related] |
-0.478 |
-0.028 |
| 104 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
-0.450 |
-0.104 |
| 105 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.431 |
0.005 |
| 106 |
262421_at |
AT1G50290
|
unknown |
-0.431 |
-0.009 |
| 107 |
254566_at |
AT4G19240
|
unknown |
-0.422 |
0.009 |
| 108 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.421 |
0.089 |
| 109 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.419 |
0.427 |
| 110 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.418 |
0.091 |
| 111 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.409 |
0.533 |
| 112 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.404 |
-0.049 |
| 113 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.401 |
0.055 |
| 114 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.390 |
0.021 |
| 115 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
-0.389 |
0.002 |
| 116 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.383 |
0.931 |
| 117 |
262452_at |
AT1G11210
|
unknown |
-0.374 |
0.762 |
| 118 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.371 |
0.210 |
| 119 |
259962_at |
AT1G53690
|
[DNA directed RNA polymerase, 7 kDa subunit] |
-0.371 |
-0.060 |
| 120 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.365 |
-0.037 |
| 121 |
246888_at |
AT5G26270
|
unknown |
-0.354 |
0.273 |
| 122 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.349 |
0.239 |
| 123 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.349 |
-0.182 |
| 124 |
253796_at |
AT4G28460
|
unknown |
-0.343 |
0.082 |
| 125 |
257817_at |
AT3G25150
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
-0.343 |
-0.031 |
| 126 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.331 |
-0.348 |
| 127 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.324 |
-0.277 |
| 128 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.318 |
0.667 |
| 129 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.315 |
0.040 |
| 130 |
254683_at |
AT4G13800
|
unknown |
-0.315 |
0.221 |
| 131 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.313 |
-0.042 |
| 132 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.312 |
-0.032 |
| 133 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
-0.305 |
0.187 |
| 134 |
259850_at |
AT1G72240
|
unknown |
-0.303 |
0.165 |
| 135 |
257925_at |
AT3G23170
|
unknown |
-0.295 |
0.469 |
| 136 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.295 |
0.399 |
| 137 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
-0.295 |
-0.082 |
| 138 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
-0.292 |
-0.107 |
| 139 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.282 |
0.701 |
| 140 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.280 |
-0.103 |
| 141 |
261374_at |
AT1G52990
|
[thioredoxin family protein] |
-0.276 |
-0.025 |
| 142 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.275 |
1.122 |
| 143 |
265539_at |
AT2G15830
|
unknown |
-0.267 |
0.140 |
| 144 |
253322_at |
AT4G33980
|
unknown |
-0.263 |
0.060 |
| 145 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
-0.260 |
0.027 |
| 146 |
249903_at |
AT5G22690
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.258 |
0.238 |
| 147 |
258351_at |
AT3G17700
|
ATCNGC20, CYCLIC NUCLEOTIDE-GATED CHANNEL 20, CNBT1, cyclic nucleotide-binding transporter 1, CNGC20, cyclic nucleotide gated channel 20 |
-0.257 |
0.151 |
| 148 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.254 |
0.042 |
| 149 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.252 |
-0.096 |
| 150 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.252 |
-0.025 |
| 151 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.245 |
0.013 |
| 152 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.239 |
-0.125 |
| 153 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
-0.237 |
-0.166 |
| 154 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.237 |
0.509 |
| 155 |
259472_at |
AT1G18910
|
[zinc ion binding] |
-0.237 |
-0.079 |
| 156 |
259250_at |
AT3G07580
|
unknown |
-0.235 |
0.147 |
| 157 |
252551_at |
AT3G45880
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.234 |
0.006 |
| 158 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.234 |
-0.010 |
| 159 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.233 |
0.090 |
| 160 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.233 |
0.167 |
| 161 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.233 |
0.086 |
| 162 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.232 |
0.213 |
| 163 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.232 |
0.029 |
| 164 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.231 |
0.498 |
| 165 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.231 |
-0.079 |
| 166 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.229 |
-0.048 |
| 167 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.229 |
-0.016 |
| 168 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.227 |
-0.365 |
| 169 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.225 |
0.067 |
| 170 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.220 |
0.387 |
| 171 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.220 |
-0.132 |
| 172 |
259683_at |
AT1G63050
|
AtLPLAT2, LPCAT2, lysophosphatidylcholine acyltransferase 2, LPLAT2, lysophospholipid acyltransferase 2 |
-0.219 |
-0.072 |
| 173 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.219 |
-0.019 |
| 174 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
-0.219 |
-0.067 |
| 175 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.218 |
-0.403 |
| 176 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.218 |
0.016 |
| 177 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.217 |
0.053 |
| 178 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.216 |
0.113 |
| 179 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.216 |
0.669 |
| 180 |
256499_at |
AT1G36640
|
unknown |
-0.215 |
0.084 |
| 181 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
-0.215 |
0.074 |