|
probeID |
AGICode |
Annotation |
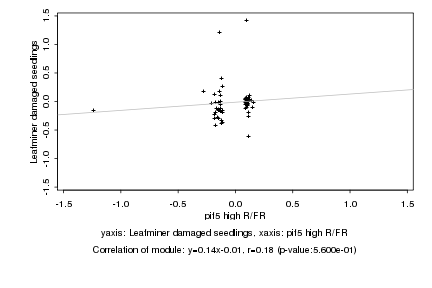
Log2 signal ratio
pif5 high R/FR |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
263260_at |
AT1G10570
|
OTS2, OVERLY TOLERANT TO SALT 2, ULP1C, UB-LIKE PROTEASE 1C |
0.154 |
-0.012 |
| 2 |
256620_at |
AT3G24480
|
[Leucine-rich repeat (LRR) family protein] |
0.144 |
-0.100 |
| 3 |
246703_at |
AT5G28080
|
WNK9 |
0.140 |
0.032 |
| 4 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.122 |
0.108 |
| 5 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
0.118 |
0.039 |
| 6 |
255730_at |
AT1G25460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.111 |
0.013 |
| 7 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.111 |
-0.247 |
| 8 |
245429_at |
AT4G17580
|
[Bax inhibitor-1 family protein] |
0.110 |
0.071 |
| 9 |
256096_at |
AT1G13650
|
unknown |
0.110 |
-0.608 |
| 10 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.110 |
-0.181 |
| 11 |
251863_at |
AT3G54870
|
ARK1, ARMADILLO REPEAT-CONTAINING KINESIN 1, AtKINUc, Arabidopsis thaliana KINESIN Ungrouped clade, gene A, CAE1, CA-ROP2 ENHANCER 1, MRH2, MORPHOGENESIS OF ROOT HAIR 2 |
0.105 |
-0.027 |
| 12 |
252709_at |
AT3G43840
|
[3-oxo-5-alpha-steroid 4-dehydrogenase family protein] |
0.103 |
0.048 |
| 13 |
258504_at |
AT3G02590
|
[Fatty acid hydroxylase superfamily protein] |
0.100 |
-0.056 |
| 14 |
249744_at |
AT5G24550
|
BGLU32, beta glucosidase 32 |
0.099 |
-0.025 |
| 15 |
262893_at |
AT1G59790
|
[Cullin family protein] |
0.097 |
-0.036 |
| 16 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.094 |
1.437 |
| 17 |
262279_at |
AT1G68630
|
[PLAC8 family protein] |
0.093 |
0.018 |
| 18 |
247088_at |
AT5G66340
|
unknown |
0.090 |
-0.026 |
| 19 |
257424_at |
AT1G78840
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.089 |
0.000 |
| 20 |
251381_at |
AT3G60720
|
PDLP8, plasmodesmata-located protein 8 |
0.089 |
-0.045 |
| 21 |
264129_at |
AT1G79170
|
unknown |
0.089 |
-0.091 |
| 22 |
256742_at |
AT3G29380
|
pBRP2, plant-specific TFIIB-related protein 2 |
0.088 |
-0.022 |
| 23 |
248897_at |
AT5G46360
|
ATKCO3, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 3, KCO3, Ca2+ activated outward rectifying K+ channel 3 |
0.088 |
0.081 |
| 24 |
256951_at |
AT3G19085
|
unknown |
0.087 |
-0.108 |
| 25 |
265973_at |
AT2G11240
|
[non-LTR retrotransposon family (LINE), has a 1.1e-38 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
0.087 |
-0.039 |
| 26 |
246526_at |
AT5G15720
|
GLIP7, GDSL-motif lipase 7 |
0.087 |
0.048 |
| 27 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
0.087 |
0.055 |
| 28 |
264541_at |
AT1G55660
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
0.086 |
0.052 |
| 29 |
253713_at |
AT4G29370
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.086 |
-0.013 |
| 30 |
264973_at |
AT1G27040
|
AtNPF4.5, NPF4.5, NRT1/ PTR family 4.5 |
0.084 |
0.009 |
| 31 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-1.240 |
-0.154 |
| 32 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.283 |
0.189 |
| 33 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.214 |
-0.034 |
| 34 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.188 |
-0.292 |
| 35 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.188 |
0.128 |
| 36 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.184 |
-0.214 |
| 37 |
252765_at |
AT3G42800
|
unknown |
-0.181 |
-0.176 |
| 38 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.180 |
-0.009 |
| 39 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.178 |
-0.416 |
| 40 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.168 |
-0.107 |
| 41 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.158 |
-0.278 |
| 42 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.154 |
-0.016 |
| 43 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.151 |
-0.287 |
| 44 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.148 |
-0.129 |
| 45 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.148 |
-0.149 |
| 46 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.147 |
1.222 |
| 47 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.144 |
0.184 |
| 48 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.134 |
0.015 |
| 49 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.133 |
-0.112 |
| 50 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.133 |
-0.174 |
| 51 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.131 |
0.122 |
| 52 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.131 |
-0.044 |
| 53 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.130 |
0.416 |
| 54 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.128 |
-0.330 |
| 55 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.127 |
-0.381 |
| 56 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.121 |
-0.156 |
| 57 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.120 |
-0.190 |
| 58 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.118 |
-0.351 |
| 59 |
246888_at |
AT5G26270
|
unknown |
-0.118 |
0.273 |