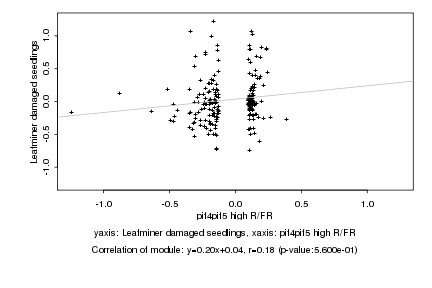
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pif4pif5 high R/FR |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.384 |
-0.270 |
| 2 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.261 |
-0.236 |
| 3 |
256789_at |
AT3G13672
|
SINA2 |
0.243 |
0.452 |
| 4 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.230 |
0.806 |
| 5 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.228 |
0.812 |
| 6 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.207 |
0.251 |
| 7 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.206 |
-0.258 |
| 8 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.196 |
0.013 |
| 9 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.196 |
0.826 |
| 10 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.187 |
0.685 |
| 11 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.187 |
0.382 |
| 12 |
256096_at |
AT1G13650
|
unknown |
0.178 |
-0.608 |
| 13 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.178 |
0.353 |
| 14 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.172 |
-0.232 |
| 15 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.162 |
0.359 |
| 16 |
251050_at |
AT5G02440
|
unknown |
0.159 |
-0.186 |
| 17 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.156 |
0.690 |
| 18 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.154 |
-0.017 |
| 19 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.152 |
0.407 |
| 20 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.150 |
0.482 |
| 21 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.146 |
-0.192 |
| 22 |
258327_at |
AT3G22640
|
PAP85 |
0.144 |
-0.038 |
| 23 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
0.143 |
0.225 |
| 24 |
246698_at |
AT5G30480
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.2e-37 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.142 |
-0.019 |
| 25 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
0.139 |
0.263 |
| 26 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.139 |
-0.481 |
| 27 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.137 |
-0.054 |
| 28 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.136 |
-0.264 |
| 29 |
263260_at |
AT1G10570
|
OTS2, OVERLY TOLERANT TO SALT 2, ULP1C, UB-LIKE PROTEASE 1C |
0.135 |
-0.012 |
| 30 |
249150_at |
AT5G43340
|
PHT1;6, phosphate transporter 1;6, PHT6, PHOSPHATE TRANSPORTER 6 |
0.133 |
0.001 |
| 31 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.133 |
0.172 |
| 32 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.132 |
-0.212 |
| 33 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.131 |
-0.402 |
| 34 |
260129_at |
AT1G36380
|
unknown |
0.130 |
0.094 |
| 35 |
251462_at |
AT3G59790
|
ATMPK10, MAP kinase 10, MPK10, MAP kinase 10 |
0.129 |
0.013 |
| 36 |
256083_at |
AT1G20735
|
[F-box and associated interaction domains-containing protein] |
0.129 |
-0.010 |
| 37 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.128 |
-0.209 |
| 38 |
262009_at |
AT1G35610
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.128 |
0.016 |
| 39 |
252298_at |
AT3G49060
|
[U-box domain-containing protein kinase family protein] |
0.127 |
-0.107 |
| 40 |
251957_at |
AT3G53690
|
[RING/U-box superfamily protein] |
0.126 |
0.058 |
| 41 |
254619_at |
AT4G18770
|
AtMYB98, myb domain protein 98, MYB98, myb domain protein 98 |
0.126 |
-0.132 |
| 42 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.126 |
0.398 |
| 43 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
0.126 |
0.016 |
| 44 |
256014_at |
AT1G19200
|
unknown |
0.125 |
0.126 |
| 45 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.124 |
0.224 |
| 46 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.123 |
1.035 |
| 47 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.122 |
0.238 |
| 48 |
261093_at |
AT1G62890
|
unknown |
0.122 |
-0.009 |
| 49 |
255295_at |
AT4G04760
|
[Major facilitator superfamily protein] |
0.122 |
-0.047 |
| 50 |
259517_at |
AT1G20630
|
CAT1, catalase 1 |
0.121 |
0.201 |
| 51 |
257424_at |
AT1G78840
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.120 |
0.000 |
| 52 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.119 |
0.087 |
| 53 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.119 |
1.077 |
| 54 |
253150_at |
AT4G35660
|
unknown |
0.119 |
0.022 |
| 55 |
264825_at |
AT1G03720
|
[Cysteine proteinases superfamily protein] |
0.116 |
-0.007 |
| 56 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
0.116 |
-0.092 |
| 57 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
0.115 |
-0.002 |
| 58 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
0.114 |
-0.036 |
| 59 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.114 |
0.008 |
| 60 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.113 |
-0.185 |
| 61 |
245429_at |
AT4G17580
|
[Bax inhibitor-1 family protein] |
0.112 |
0.071 |
| 62 |
257578_x_at |
AT3G30160
|
unknown |
0.111 |
0.047 |
| 63 |
267135_at |
AT2G23430
|
ICK1, KRP1, KIP-RELATED PROTEIN 1 |
0.110 |
-0.195 |
| 64 |
261162_at |
AT1G34440
|
unknown |
0.110 |
-0.008 |
| 65 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.110 |
-0.050 |
| 66 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.110 |
0.175 |
| 67 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.110 |
-0.187 |
| 68 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.110 |
0.605 |
| 69 |
249902_at |
AT5G22680
|
unknown |
0.109 |
0.016 |
| 70 |
251445_at |
AT3G59870
|
unknown |
0.109 |
0.034 |
| 71 |
261908_at |
AT1G50650
|
[Stigma-specific Stig1 family protein] |
0.109 |
0.032 |
| 72 |
263625_at |
AT2G04920
|
[F-box and associated interaction domains-containing protein] |
0.109 |
0.042 |
| 73 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
0.109 |
-0.488 |
| 74 |
251381_at |
AT3G60720
|
PDLP8, plasmodesmata-located protein 8 |
0.109 |
-0.045 |
| 75 |
259635_at |
AT1G56360
|
ATPAP6, PAP6, purple acid phosphatase 6 |
0.109 |
0.059 |
| 76 |
248026_at |
AT5G55710
|
AtTic20-V, translocon at the inner envelope membrane of chloroplasts 20-V, Tic20-V, translocon at the inner envelope membrane of chloroplasts 20-V |
0.108 |
-0.081 |
| 77 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.108 |
-0.398 |
| 78 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.107 |
0.807 |
| 79 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.106 |
0.793 |
| 80 |
257230_at |
AT3G16540
|
DEG11, degradation of periplasmic proteins 11, DegP11, DegP protease 11 |
0.106 |
-0.018 |
| 81 |
258358_at |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
0.106 |
0.064 |
| 82 |
246524_at |
AT5G15860
|
ATPCME, prenylcysteine methylesterase, ICME, Isoprenylcysteine methylesterase, PCME, prenylcysteine methylesterase |
0.105 |
-0.263 |
| 83 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.105 |
0.035 |
| 84 |
246874_at |
AT5G26120
|
ASD2, alpha-L-arabinofuranosidase 2, ATASD2, ARABIDOPSIS THALIANA ALPHA-L-ARABINOFURANOSIDASE 2 |
0.105 |
-0.024 |
| 85 |
262927_at |
AT1G65810
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.104 |
-0.001 |
| 86 |
248586_at |
AT5G49610
|
[F-box family protein] |
0.104 |
-0.056 |
| 87 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.104 |
0.090 |
| 88 |
262566_at |
AT1G34310
|
ARF12, auxin response factor 12 |
0.104 |
-0.204 |
| 89 |
256371_at |
AT1G66770
|
AtSWEET6, SWEET6 |
0.104 |
-0.045 |
| 90 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.103 |
-0.112 |
| 91 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.103 |
0.000 |
| 92 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.103 |
-0.733 |
| 93 |
266658_at |
AT2G25735
|
unknown |
0.103 |
0.857 |
| 94 |
251302_at |
AT3G61970
|
NGA2, NGATHA2 |
0.102 |
-0.205 |
| 95 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.102 |
-0.412 |
| 96 |
252455_at |
AT3G47140
|
[F-box associated ubiquitination effector family protein] |
0.101 |
0.027 |
| 97 |
252301_at |
AT3G49162
|
|
0.101 |
-0.031 |
| 98 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.101 |
-0.100 |
| 99 |
249114_at |
AT5G43800
|
[copia-like retrotransposon family, has a 0. P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.101 |
-0.063 |
| 100 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
0.101 |
-0.052 |
| 101 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.101 |
-0.132 |
| 102 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.100 |
0.098 |
| 103 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.100 |
0.441 |
| 104 |
245661_at |
AT1G28220
|
ATPUP3, purine permease 3, PUP3, purine permease 3 |
0.099 |
-0.044 |
| 105 |
258520_at |
AT3G06710
|
unknown |
0.099 |
-0.031 |
| 106 |
257317_at |
ATMG01060
|
ORF107G |
0.099 |
0.047 |
| 107 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
0.099 |
0.022 |
| 108 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.098 |
0.653 |
| 109 |
257870_at |
AT3G28490
|
[Oxoglutarate/iron-dependent oxygenase] |
0.097 |
0.038 |
| 110 |
256708_at |
AT3G30320
|
unknown |
0.097 |
0.019 |
| 111 |
257588_x_at |
AT1G30150
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 2.2e-24 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
0.097 |
-0.022 |
| 112 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.097 |
-0.423 |
| 113 |
245185_at |
AT1G67760
|
[TCP-1/cpn60 chaperonin family protein] |
0.096 |
-0.032 |
| 114 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-1.249 |
-0.154 |
| 115 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.882 |
0.128 |
| 116 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.644 |
-0.138 |
| 117 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.517 |
0.184 |
| 118 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.499 |
-0.284 |
| 119 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.473 |
-0.034 |
| 120 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.472 |
-0.292 |
| 121 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.466 |
-0.214 |
| 122 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.440 |
-0.129 |
| 123 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.355 |
-0.381 |
| 124 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.354 |
0.189 |
| 125 |
252765_at |
AT3G42800
|
unknown |
-0.351 |
-0.176 |
| 126 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.347 |
1.073 |
| 127 |
258196_at |
AT3G13980
|
unknown |
-0.344 |
-0.156 |
| 128 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.329 |
-0.416 |
| 129 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.320 |
-0.330 |
| 130 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.318 |
-0.311 |
| 131 |
263002_at |
AT1G54200
|
unknown |
-0.317 |
-0.530 |
| 132 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.316 |
0.545 |
| 133 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.313 |
-0.002 |
| 134 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.306 |
0.689 |
| 135 |
250327_at |
AT5G12050
|
unknown |
-0.304 |
-0.252 |
| 136 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.303 |
-0.187 |
| 137 |
248509_at |
AT5G50335
|
unknown |
-0.295 |
0.063 |
| 138 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.282 |
-0.154 |
| 139 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.282 |
0.000 |
| 140 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.280 |
0.122 |
| 141 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.272 |
-0.276 |
| 142 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.269 |
0.322 |
| 143 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.268 |
-0.365 |
| 144 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.261 |
-0.112 |
| 145 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.246 |
0.107 |
| 146 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.246 |
-0.035 |
| 147 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.240 |
-0.009 |
| 148 |
247878_at |
AT5G57760
|
unknown |
-0.237 |
-0.372 |
| 149 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.237 |
-0.093 |
| 150 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.234 |
0.213 |
| 151 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.232 |
-0.278 |
| 152 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.231 |
0.758 |
| 153 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.230 |
0.052 |
| 154 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.229 |
-0.040 |
| 155 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.228 |
0.727 |
| 156 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.221 |
-0.031 |
| 157 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.220 |
-0.404 |
| 158 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.211 |
0.141 |
| 159 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.210 |
-0.496 |
| 160 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.205 |
0.283 |
| 161 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.204 |
0.158 |
| 162 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.204 |
-0.032 |
| 163 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.202 |
-0.120 |
| 164 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.202 |
0.288 |
| 165 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.201 |
-0.293 |
| 166 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
-0.199 |
0.020 |
| 167 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.198 |
0.000 |
| 168 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.197 |
-0.203 |
| 169 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.196 |
-0.327 |
| 170 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.196 |
0.041 |
| 171 |
261684_at |
AT1G47400
|
unknown |
-0.194 |
-0.436 |
| 172 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.194 |
-0.342 |
| 173 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.193 |
-0.004 |
| 174 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.188 |
0.285 |
| 175 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.183 |
0.340 |
| 176 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.182 |
1.005 |
| 177 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.181 |
-0.208 |
| 178 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.179 |
-0.016 |
| 179 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.176 |
-0.341 |
| 180 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.176 |
-0.133 |
| 181 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.173 |
-0.176 |
| 182 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.173 |
1.222 |
| 183 |
250937_at |
AT5G03230
|
unknown |
-0.170 |
0.322 |
| 184 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.169 |
-0.491 |
| 185 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.169 |
0.193 |
| 186 |
256617_at |
AT3G22240
|
unknown |
-0.164 |
-0.079 |
| 187 |
259884_at |
AT1G76390
|
PUB43, plant U-box 43 |
-0.163 |
0.398 |
| 188 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.162 |
-0.174 |
| 189 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.162 |
-0.227 |
| 190 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.161 |
0.046 |
| 191 |
265611_at |
AT2G25510
|
unknown |
-0.159 |
-0.009 |
| 192 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.155 |
-0.019 |
| 193 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.155 |
-0.407 |
| 194 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.154 |
0.030 |
| 195 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.154 |
-0.499 |
| 196 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.154 |
-0.351 |
| 197 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.153 |
0.113 |
| 198 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.152 |
0.140 |
| 199 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.151 |
-0.213 |
| 200 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.150 |
-0.003 |
| 201 |
257875_at |
AT3G17120
|
unknown |
-0.150 |
-0.001 |
| 202 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
-0.149 |
-0.303 |
| 203 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.148 |
0.188 |
| 204 |
246888_at |
AT5G26270
|
unknown |
-0.148 |
0.273 |
| 205 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.147 |
-0.503 |
| 206 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.147 |
-0.266 |
| 207 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.147 |
0.090 |
| 208 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.145 |
-0.712 |
| 209 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
-0.144 |
0.197 |
| 210 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.144 |
-0.720 |
| 211 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.143 |
-0.171 |
| 212 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.143 |
-0.240 |
| 213 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.143 |
-0.107 |
| 214 |
245076_at |
AT2G23170
|
GH3.3 |
-0.141 |
0.781 |
| 215 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.139 |
0.169 |
| 216 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
-0.139 |
0.865 |
| 217 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.136 |
-0.180 |
| 218 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.135 |
0.072 |
| 219 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.134 |
-0.171 |
| 220 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.133 |
0.635 |
| 221 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
-0.133 |
-0.070 |
| 222 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.131 |
0.120 |
| 223 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.131 |
0.464 |
| 224 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.130 |
-0.124 |
| 225 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.130 |
-0.147 |