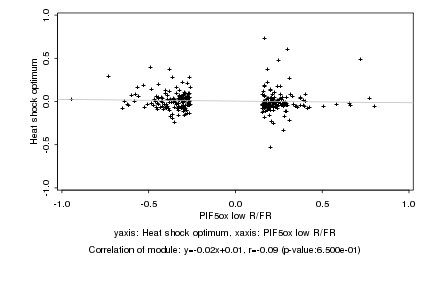
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PIF5ox low R/FR |
Log2 signal ratio
Heat shock optimum |
| 1 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.801 |
-0.056 |
| 2 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.769 |
0.037 |
| 3 |
261684_at |
AT1G47400
|
unknown |
0.716 |
0.496 |
| 4 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.658 |
-0.045 |
| 5 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.656 |
-0.020 |
| 6 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.578 |
-0.023 |
| 7 |
245632_at |
AT1G25290
|
ATRBL10, RHOMBOID-like protein 10, ATRBL7, RHOMBOID-like protein 7, AtRBL8, RBL10, RHOMBOID-like protein 10, RBL8, RHOMBOID-like protein 8 |
0.502 |
-0.048 |
| 8 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.425 |
-0.058 |
| 9 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.411 |
-0.076 |
| 10 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.402 |
0.088 |
| 11 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.398 |
0.008 |
| 12 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.397 |
-0.051 |
| 13 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.392 |
0.014 |
| 14 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.387 |
-0.042 |
| 15 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.374 |
-0.040 |
| 16 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.374 |
0.054 |
| 17 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.370 |
0.043 |
| 18 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.355 |
-0.066 |
| 19 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.351 |
-0.050 |
| 20 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.341 |
-0.054 |
| 21 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.330 |
-0.027 |
| 22 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.327 |
0.059 |
| 23 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.315 |
0.082 |
| 24 |
259001_at |
AT3G01960
|
unknown |
0.311 |
-0.210 |
| 25 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.307 |
0.271 |
| 26 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.297 |
0.610 |
| 27 |
263227_at |
AT1G30750
|
unknown |
0.296 |
-0.044 |
| 28 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.293 |
-0.105 |
| 29 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.291 |
0.058 |
| 30 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.290 |
-0.037 |
| 31 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
0.290 |
-0.015 |
| 32 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.284 |
-0.010 |
| 33 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.283 |
-0.111 |
| 34 |
258807_at |
AT3G04030
|
MYR2 |
0.281 |
-0.167 |
| 35 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
0.279 |
-0.033 |
| 36 |
249614_at |
AT5G37300
|
WSD1 |
0.276 |
-0.329 |
| 37 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.275 |
-0.048 |
| 38 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.275 |
0.001 |
| 39 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
0.271 |
-0.016 |
| 40 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.271 |
0.047 |
| 41 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.263 |
0.025 |
| 42 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.262 |
-0.040 |
| 43 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.258 |
0.036 |
| 44 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.257 |
0.083 |
| 45 |
250828_at |
AT5G05250
|
unknown |
0.256 |
0.181 |
| 46 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.254 |
-0.014 |
| 47 |
245328_at |
AT4G14465
|
AHL20, AT-hook motif nuclear-localized protein 20 |
0.251 |
0.029 |
| 48 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.248 |
-0.035 |
| 49 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.244 |
0.482 |
| 50 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.244 |
-0.005 |
| 51 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.242 |
0.174 |
| 52 |
247553_at |
AT5G60910
|
AGL8, AGAMOUS-like 8, FUL, FRUITFULL |
0.241 |
-0.071 |
| 53 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.239 |
0.020 |
| 54 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.239 |
-0.052 |
| 55 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.235 |
0.009 |
| 56 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.233 |
-0.023 |
| 57 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.228 |
-0.065 |
| 58 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.227 |
-0.009 |
| 59 |
263829_at |
AT2G40435
|
unknown |
0.227 |
-0.024 |
| 60 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.224 |
0.107 |
| 61 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.222 |
0.048 |
| 62 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.222 |
-0.005 |
| 63 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
0.219 |
0.032 |
| 64 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.219 |
0.009 |
| 65 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
0.218 |
-0.006 |
| 66 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.217 |
-0.059 |
| 67 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.217 |
-0.015 |
| 68 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
0.216 |
-0.027 |
| 69 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.216 |
-0.253 |
| 70 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.215 |
-0.096 |
| 71 |
249367_at |
AT5G40630
|
[Ubiquitin-like superfamily protein] |
0.214 |
-0.048 |
| 72 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.213 |
-0.055 |
| 73 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.211 |
0.032 |
| 74 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.210 |
-0.041 |
| 75 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.208 |
0.084 |
| 76 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.207 |
0.050 |
| 77 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.207 |
-0.225 |
| 78 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
0.204 |
0.035 |
| 79 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
0.203 |
0.009 |
| 80 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.202 |
0.003 |
| 81 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.202 |
-0.100 |
| 82 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.202 |
0.042 |
| 83 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
0.201 |
0.131 |
| 84 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.200 |
-0.009 |
| 85 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.200 |
0.143 |
| 86 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.199 |
-0.528 |
| 87 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.199 |
-0.063 |
| 88 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
0.197 |
-0.103 |
| 89 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.196 |
-0.151 |
| 90 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
0.194 |
-0.044 |
| 91 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
0.193 |
-0.033 |
| 92 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.189 |
0.005 |
| 93 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
0.189 |
0.039 |
| 94 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
0.188 |
-0.020 |
| 95 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.188 |
-0.086 |
| 96 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
0.186 |
-0.074 |
| 97 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.186 |
-0.043 |
| 98 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.184 |
0.225 |
| 99 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.184 |
0.040 |
| 100 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.183 |
-0.069 |
| 101 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.183 |
0.377 |
| 102 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.183 |
-0.066 |
| 103 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
0.181 |
-0.099 |
| 104 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.178 |
-0.045 |
| 105 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.178 |
-0.044 |
| 106 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
0.178 |
-0.041 |
| 107 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
0.177 |
0.032 |
| 108 |
255422_at |
AT4G03270
|
CYCD6;1, Cyclin D6;1 |
0.176 |
-0.051 |
| 109 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.174 |
-0.029 |
| 110 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.174 |
0.008 |
| 111 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.172 |
-0.005 |
| 112 |
254043_at |
AT4G25990
|
CIL |
0.172 |
-0.033 |
| 113 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.170 |
-0.113 |
| 114 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.169 |
-0.054 |
| 115 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.167 |
-0.046 |
| 116 |
265935_at |
AT2G19580
|
TET2, tetraspanin2 |
0.167 |
-0.029 |
| 117 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
0.167 |
-0.050 |
| 118 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.167 |
0.060 |
| 119 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.166 |
0.175 |
| 120 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
0.166 |
-0.038 |
| 121 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.166 |
-0.175 |
| 122 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.165 |
0.732 |
| 123 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
0.164 |
0.004 |
| 124 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.164 |
0.196 |
| 125 |
247246_at |
AT5G64620
|
ATC/VIF2, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 2, C/VIF2, cell wall / vacuolar inhibitor of fructosidase 2 |
0.164 |
0.076 |
| 126 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
0.164 |
-0.074 |
| 127 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.162 |
-0.061 |
| 128 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
0.161 |
-0.030 |
| 129 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.161 |
-0.112 |
| 130 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.160 |
0.127 |
| 131 |
248590_at |
AT5G49660
|
XIP1, XYLEM INTERMIXED WITH PHLOEM 1 |
0.160 |
-0.046 |
| 132 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
0.158 |
0.007 |
| 133 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.156 |
-0.014 |
| 134 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.155 |
-0.031 |
| 135 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.155 |
0.082 |
| 136 |
263632_at |
AT2G04795
|
unknown |
0.155 |
-0.125 |
| 137 |
264068_at |
AT2G27990
|
BLH8, BEL1-like homeodomain 8, PNF, POUND-FOOLISH |
0.155 |
-0.027 |
| 138 |
258738_at |
AT3G05750
|
TRM6, TON1 Recruiting Motif 6 |
0.154 |
-0.072 |
| 139 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.154 |
-0.043 |
| 140 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.949 |
0.032 |
| 141 |
245076_at |
AT2G23170
|
GH3.3 |
-0.735 |
0.299 |
| 142 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.654 |
-0.071 |
| 143 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.645 |
0.006 |
| 144 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.625 |
-0.025 |
| 145 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.617 |
-0.040 |
| 146 |
248509_at |
AT5G50335
|
unknown |
-0.604 |
0.078 |
| 147 |
266222_at |
AT2G28780
|
unknown |
-0.584 |
0.006 |
| 148 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.581 |
0.086 |
| 149 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.566 |
0.165 |
| 150 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.560 |
0.064 |
| 151 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.535 |
0.189 |
| 152 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.529 |
-0.059 |
| 153 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.511 |
-0.024 |
| 154 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.495 |
0.395 |
| 155 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.486 |
0.140 |
| 156 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.485 |
-0.017 |
| 157 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.477 |
-0.039 |
| 158 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.467 |
0.026 |
| 159 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.461 |
0.007 |
| 160 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.459 |
0.068 |
| 161 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.456 |
-0.063 |
| 162 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.453 |
-0.083 |
| 163 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.452 |
-0.021 |
| 164 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.448 |
0.045 |
| 165 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.448 |
-0.031 |
| 166 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.447 |
0.199 |
| 167 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.446 |
0.055 |
| 168 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.432 |
-0.061 |
| 169 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.430 |
0.006 |
| 170 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
-0.428 |
0.048 |
| 171 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.426 |
-0.050 |
| 172 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.426 |
-0.009 |
| 173 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.425 |
0.046 |
| 174 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.419 |
-0.081 |
| 175 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.414 |
-0.028 |
| 176 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.407 |
0.130 |
| 177 |
247474_at |
AT5G62280
|
unknown |
-0.404 |
-0.069 |
| 178 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.404 |
0.094 |
| 179 |
248801_at |
AT5G47370
|
HAT2 |
-0.399 |
-0.024 |
| 180 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.399 |
-0.072 |
| 181 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.397 |
0.075 |
| 182 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.396 |
0.033 |
| 183 |
250438_at |
AT5G10580
|
unknown |
-0.393 |
-0.055 |
| 184 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.387 |
-0.075 |
| 185 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.385 |
0.016 |
| 186 |
261292_at |
AT1G36940
|
unknown |
-0.384 |
0.116 |
| 187 |
265913_at |
AT2G25625
|
unknown |
-0.384 |
-0.008 |
| 188 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.383 |
0.371 |
| 189 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.382 |
-0.102 |
| 190 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.379 |
0.028 |
| 191 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.375 |
-0.169 |
| 192 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.374 |
-0.000 |
| 193 |
252204_at |
AT3G50340
|
unknown |
-0.368 |
-0.147 |
| 194 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.364 |
0.284 |
| 195 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.363 |
-0.187 |
| 196 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.362 |
0.027 |
| 197 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.358 |
-0.007 |
| 198 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.358 |
0.039 |
| 199 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.355 |
0.006 |
| 200 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.355 |
-0.064 |
| 201 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.355 |
-0.238 |
| 202 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.352 |
0.028 |
| 203 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.349 |
0.000 |
| 204 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.343 |
-0.034 |
| 205 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.341 |
0.104 |
| 206 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.340 |
0.164 |
| 207 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.334 |
-0.072 |
| 208 |
267503_at |
AT2G45600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.333 |
-0.100 |
| 209 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.333 |
-0.056 |
| 210 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.331 |
-0.019 |
| 211 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.330 |
-0.157 |
| 212 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.329 |
0.044 |
| 213 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.329 |
-0.019 |
| 214 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.328 |
-0.065 |
| 215 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.327 |
0.033 |
| 216 |
252765_at |
AT3G42800
|
unknown |
-0.326 |
0.073 |
| 217 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.326 |
0.015 |
| 218 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.325 |
0.134 |
| 219 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.325 |
0.062 |
| 220 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.320 |
0.049 |
| 221 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.316 |
-0.050 |
| 222 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.315 |
0.017 |
| 223 |
265276_at |
AT2G28400
|
unknown |
-0.315 |
0.060 |
| 224 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.313 |
0.038 |
| 225 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.313 |
-0.080 |
| 226 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
-0.310 |
0.069 |
| 227 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.309 |
-0.049 |
| 228 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.308 |
0.038 |
| 229 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.307 |
0.000 |
| 230 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.306 |
0.222 |
| 231 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.305 |
-0.000 |
| 232 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.305 |
-0.111 |
| 233 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.304 |
0.042 |
| 234 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.303 |
-0.074 |
| 235 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.302 |
-0.035 |
| 236 |
254758_at |
AT4G13260
|
AtYUC2, YUC2, YUCCA2 |
-0.302 |
-0.066 |
| 237 |
263931_at |
AT2G36220
|
unknown |
-0.302 |
0.087 |
| 238 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.301 |
0.074 |
| 239 |
249136_at |
AT5G43180
|
unknown |
-0.301 |
-0.062 |
| 240 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
-0.300 |
-0.037 |
| 241 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.298 |
0.110 |
| 242 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.298 |
0.001 |
| 243 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.298 |
-0.151 |
| 244 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.297 |
-0.091 |
| 245 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.296 |
-0.060 |
| 246 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.296 |
-0.078 |
| 247 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.292 |
-0.140 |
| 248 |
263002_at |
AT1G54200
|
unknown |
-0.292 |
0.101 |
| 249 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.291 |
0.051 |
| 250 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.291 |
-0.076 |
| 251 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
-0.289 |
-0.043 |
| 252 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.286 |
-0.099 |
| 253 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.285 |
0.057 |
| 254 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.285 |
0.026 |
| 255 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.283 |
-0.097 |
| 256 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.282 |
-0.131 |
| 257 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.281 |
0.219 |
| 258 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.280 |
-0.043 |
| 259 |
262446_at |
AT1G49310
|
unknown |
-0.279 |
0.062 |
| 260 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.275 |
0.046 |
| 261 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.275 |
0.100 |
| 262 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.275 |
-0.028 |
| 263 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.275 |
-0.042 |
| 264 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.272 |
0.092 |
| 265 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.271 |
-0.010 |
| 266 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.271 |
-0.013 |
| 267 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
-0.270 |
0.054 |
| 268 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.269 |
0.016 |
| 269 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.269 |
0.094 |
| 270 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.268 |
-0.031 |
| 271 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.267 |
0.008 |
| 272 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.266 |
0.281 |
| 273 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.266 |
-0.136 |
| 274 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.265 |
-0.036 |
| 275 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.265 |
0.056 |
| 276 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.262 |
0.169 |
| 277 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.261 |
-0.033 |