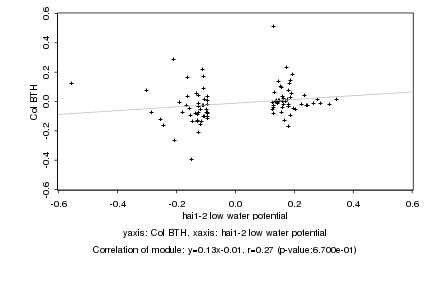
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hai1-2 low water potential |
Log2 signal ratio
Col BTH |
| 1 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.342 |
0.020 |
| 2 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.316 |
-0.017 |
| 3 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.288 |
-0.011 |
| 4 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.278 |
0.018 |
| 5 |
250826_at |
AT5G05220
|
unknown |
0.263 |
-0.012 |
| 6 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.244 |
-0.026 |
| 7 |
253494_at |
AT4G31830
|
unknown |
0.239 |
-0.021 |
| 8 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.232 |
0.044 |
| 9 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.223 |
-0.016 |
| 10 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.201 |
-0.048 |
| 11 |
245717_at |
AT5G33390
|
[glycine-rich protein] |
0.197 |
-0.046 |
| 12 |
249454_at |
AT5G39520
|
unknown |
0.191 |
0.186 |
| 13 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.189 |
0.057 |
| 14 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.186 |
0.149 |
| 15 |
254683_at |
AT4G13800
|
unknown |
0.186 |
-0.093 |
| 16 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.186 |
0.030 |
| 17 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.181 |
0.129 |
| 18 |
258248_at |
AT3G29140
|
unknown |
0.180 |
-0.032 |
| 19 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.179 |
-0.169 |
| 20 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.178 |
-0.019 |
| 21 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.178 |
0.080 |
| 22 |
249246_at |
AT5G42290
|
[transcription activator-related] |
0.175 |
0.017 |
| 23 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.172 |
0.235 |
| 24 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.169 |
0.002 |
| 25 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.164 |
-0.126 |
| 26 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.161 |
-0.018 |
| 27 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.160 |
0.024 |
| 28 |
260264_at |
AT1G68500
|
unknown |
0.159 |
0.036 |
| 29 |
255932_at |
AT1G12720
|
[Mutator-like transposase family, has a 1.2e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.159 |
0.005 |
| 30 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.157 |
-0.039 |
| 31 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.155 |
-0.074 |
| 32 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.155 |
0.102 |
| 33 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.151 |
0.104 |
| 34 |
265276_at |
AT2G28400
|
unknown |
0.149 |
0.008 |
| 35 |
246796_at |
AT5G26770
|
unknown |
0.146 |
0.137 |
| 36 |
259025_at |
AT3G33073
|
transposable element gene |
0.144 |
0.007 |
| 37 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.142 |
-0.009 |
| 38 |
254196_at |
AT4G24030
|
unknown |
0.140 |
-0.004 |
| 39 |
252665_at |
AT3G44140
|
unknown |
0.139 |
0.009 |
| 40 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
0.135 |
0.004 |
| 41 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.130 |
0.064 |
| 42 |
263881_at |
AT2G21820
|
unknown |
0.129 |
-0.039 |
| 43 |
259578_at |
AT1G27990
|
unknown |
0.128 |
-0.077 |
| 44 |
260005_at |
AT1G67920
|
unknown |
0.127 |
0.513 |
| 45 |
264966_at |
AT1G60570
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.127 |
-0.021 |
| 46 |
257776_at |
AT3G29190
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.125 |
-0.003 |
| 47 |
251572_at |
AT3G58390
|
[Eukaryotic release factor 1 (eRF1) family protein] |
0.123 |
-0.052 |
| 48 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.559 |
0.126 |
| 49 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.305 |
0.077 |
| 50 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.285 |
-0.070 |
| 51 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.255 |
-0.119 |
| 52 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.245 |
-0.161 |
| 53 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.213 |
0.293 |
| 54 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.210 |
-0.261 |
| 55 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.192 |
-0.002 |
| 56 |
255876_at |
AT2G40480
|
unknown |
-0.183 |
-0.069 |
| 57 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.168 |
-0.024 |
| 58 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.166 |
0.164 |
| 59 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.163 |
0.037 |
| 60 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.156 |
-0.042 |
| 61 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.155 |
-0.094 |
| 62 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.150 |
-0.395 |
| 63 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.147 |
-0.135 |
| 64 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.138 |
-0.081 |
| 65 |
251527_at |
AT3G58650
|
TRM7, TON1 Recruiting Motif 7 |
-0.135 |
0.056 |
| 66 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.131 |
-0.088 |
| 67 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.131 |
-0.128 |
| 68 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
-0.130 |
-0.135 |
| 69 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.128 |
-0.031 |
| 70 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.128 |
-0.209 |
| 71 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.128 |
-0.069 |
| 72 |
261794_at |
AT1G16060
|
ADAP, ARIA-interacting double AP2 domain protein, WRI3, WRINKLED 3 |
-0.128 |
0.043 |
| 73 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.126 |
-0.009 |
| 74 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.122 |
-0.152 |
| 75 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.120 |
-0.053 |
| 76 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.116 |
-0.133 |
| 77 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
-0.114 |
0.225 |
| 78 |
246505_at |
AT5G16250
|
unknown |
-0.113 |
-0.024 |
| 79 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.111 |
0.095 |
| 80 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.110 |
0.175 |
| 81 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.109 |
-0.100 |
| 82 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.109 |
-0.024 |
| 83 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.108 |
0.015 |
| 84 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.107 |
-0.102 |
| 85 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.100 |
-0.072 |
| 86 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.099 |
-0.050 |
| 87 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.098 |
-0.078 |
| 88 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.097 |
-0.019 |
| 89 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.097 |
0.039 |
| 90 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.096 |
0.010 |
| 91 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.096 |
-0.113 |
| 92 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.096 |
-0.097 |
| 93 |
254341_at |
AT4G22130
|
SRF8, STRUBBELIG-receptor family 8 |
-0.096 |
-0.073 |