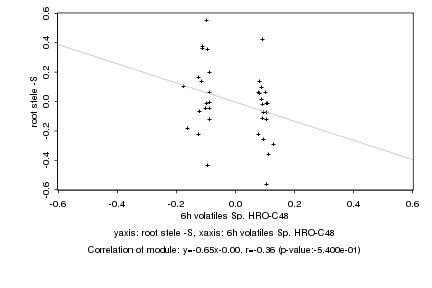
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
6h volatiles Sp. HRO-C48 |
Log2 signal ratio
root stele -S |
| 1 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.128 |
-0.288 |
| 2 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
0.110 |
-0.354 |
| 3 |
255976_at |
AT1G22010
|
unknown |
0.106 |
-0.013 |
| 4 |
247647_at |
AT5G60010
|
[ferric reductase-like transmembrane component family protein] |
0.104 |
-0.010 |
| 5 |
258608_at |
AT3G03020
|
unknown |
0.103 |
-0.558 |
| 6 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.103 |
-0.121 |
| 7 |
249731_at |
AT5G24440
|
CID13, CTC-interacting domain 13 |
0.102 |
-0.071 |
| 8 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.100 |
0.063 |
| 9 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
0.094 |
-0.252 |
| 10 |
259781_at |
AT1G29650
|
[non-LTR retrotransposon family (LINE), has a 1.5e-28 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
0.092 |
-0.070 |
| 11 |
267204_at |
AT2G31050
|
[Cupredoxin superfamily protein] |
0.091 |
-0.111 |
| 12 |
249401_at |
AT5G40260
|
AtSWEET8, RPG1, RUPTURED POLLEN GRAIN1, SWEET8 |
0.091 |
-0.014 |
| 13 |
263837_at |
AT2G04500
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.090 |
0.425 |
| 14 |
266160_at |
AT2G28180
|
ATCHX8, CHX08, CHX8, CATION/H+ EXCHANGER 8 |
0.088 |
0.019 |
| 15 |
259492_at |
AT1G15830
|
unknown |
0.086 |
0.096 |
| 16 |
251214_at |
AT3G62500
|
unknown |
0.079 |
0.143 |
| 17 |
259720_at |
AT1G61080
|
[Hydroxyproline-rich glycoprotein family protein] |
0.079 |
0.055 |
| 18 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.076 |
-0.221 |
| 19 |
265559_at |
AT2G05530
|
[Glycine-rich protein family] |
0.076 |
0.068 |
| 20 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.178 |
0.106 |
| 21 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
-0.165 |
-0.178 |
| 22 |
266164_at |
AT2G28050
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.127 |
0.169 |
| 23 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.126 |
-0.224 |
| 24 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.125 |
-0.067 |
| 25 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.117 |
0.139 |
| 26 |
265831_at |
AT2G14460
|
unknown |
-0.114 |
0.366 |
| 27 |
267309_at |
AT2G19385
|
[zinc ion binding] |
-0.113 |
0.375 |
| 28 |
252227_at |
AT3G49900
|
[Phototropic-responsive NPH3 family protein] |
-0.104 |
-0.045 |
| 29 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
-0.101 |
0.552 |
| 30 |
264310_at |
AT1G62030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.101 |
-0.009 |
| 31 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.097 |
0.356 |
| 32 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.095 |
-0.429 |
| 33 |
260108_at |
AT1G63280
|
[Serine protease inhibitor (SERPIN) family protein] |
-0.091 |
-0.047 |
| 34 |
250256_at |
AT5G13650
|
SVR3, SUPPRESSOR OF VARIEGATION 3 |
-0.090 |
0.199 |
| 35 |
266540_at |
AT2G35310
|
[Transcriptional factor B3 family protein] |
-0.090 |
-0.002 |
| 36 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.089 |
-0.119 |
| 37 |
266508_at |
AT2G47920
|
NET3C, Networked 3C |
-0.088 |
0.064 |