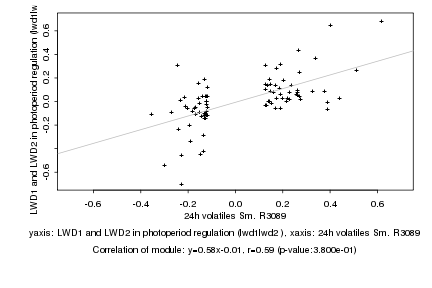
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
24h volatiles Sm. R3089 |
Log2 signal ratio
LWD1 and LWD2 in photoperiod regulation (lwd1lwd2 ) |
| 1 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.614 |
0.682 |
| 2 |
261658_at |
AT1G50040
|
unknown |
0.511 |
0.270 |
| 3 |
260495_at |
AT2G41810
|
unknown |
0.436 |
0.027 |
| 4 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.401 |
0.648 |
| 5 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.388 |
-0.004 |
| 6 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.387 |
-0.064 |
| 7 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.374 |
0.086 |
| 8 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.335 |
0.369 |
| 9 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
0.325 |
0.088 |
| 10 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
0.274 |
0.020 |
| 11 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.270 |
0.248 |
| 12 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.268 |
0.046 |
| 13 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
0.264 |
0.434 |
| 14 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.262 |
0.084 |
| 15 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.261 |
0.099 |
| 16 |
264361_at |
AT1G03300
|
unknown |
0.260 |
0.059 |
| 17 |
264521_at |
AT1G10020
|
unknown |
0.255 |
0.067 |
| 18 |
246926_at |
AT5G25240
|
unknown |
0.235 |
0.143 |
| 19 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.227 |
0.078 |
| 20 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.226 |
0.023 |
| 21 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.217 |
0.032 |
| 22 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
0.215 |
0.008 |
| 23 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.200 |
0.182 |
| 24 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.196 |
0.031 |
| 25 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.189 |
0.320 |
| 26 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.188 |
-0.055 |
| 27 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
0.187 |
0.064 |
| 28 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
0.183 |
0.117 |
| 29 |
245906_at |
AT5G11070
|
unknown |
0.172 |
0.282 |
| 30 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
0.170 |
0.027 |
| 31 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.167 |
-0.051 |
| 32 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
0.166 |
0.140 |
| 33 |
253583_at |
AT4G30680
|
[Initiation factor eIF-4 gamma, MA3] |
0.160 |
0.080 |
| 34 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.150 |
-0.015 |
| 35 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.148 |
0.089 |
| 36 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.148 |
0.151 |
| 37 |
259996_at |
AT1G67910
|
unknown |
0.143 |
0.008 |
| 38 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.142 |
0.194 |
| 39 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.139 |
0.004 |
| 40 |
258939_at |
AT3G10020
|
unknown |
0.132 |
0.143 |
| 41 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.131 |
-0.030 |
| 42 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
0.126 |
0.152 |
| 43 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.126 |
-0.031 |
| 44 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.125 |
0.310 |
| 45 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.124 |
0.108 |
| 46 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-0.355 |
-0.105 |
| 47 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.301 |
-0.536 |
| 48 |
266757_at |
AT2G46940
|
unknown |
-0.271 |
-0.088 |
| 49 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.246 |
0.313 |
| 50 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.243 |
-0.235 |
| 51 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.236 |
0.011 |
| 52 |
265892_at |
AT2G15020
|
unknown |
-0.230 |
-0.696 |
| 53 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.228 |
-0.454 |
| 54 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
-0.217 |
0.039 |
| 55 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.212 |
-0.041 |
| 56 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.206 |
-0.059 |
| 57 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.197 |
-0.202 |
| 58 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.190 |
-0.334 |
| 59 |
246236_at |
AT4G36470
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.185 |
-0.082 |
| 60 |
251944_at |
AT3G53510
|
ABCG20, ATP-binding cassette G20 |
-0.174 |
-0.055 |
| 61 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.173 |
-0.104 |
| 62 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
-0.172 |
-0.050 |
| 63 |
248959_at |
AT5G45630
|
unknown |
-0.160 |
0.153 |
| 64 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.157 |
0.031 |
| 65 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.155 |
-0.010 |
| 66 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.155 |
-0.092 |
| 67 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.151 |
-0.447 |
| 68 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.146 |
-0.123 |
| 69 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
-0.142 |
0.044 |
| 70 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.138 |
-0.288 |
| 71 |
247055_at |
AT5G66740
|
unknown |
-0.136 |
-0.416 |
| 72 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.134 |
0.190 |
| 73 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.134 |
-0.137 |
| 74 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
-0.132 |
-0.112 |
| 75 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
-0.132 |
-0.095 |
| 76 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.130 |
-0.138 |
| 77 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.129 |
-0.095 |
| 78 |
246125_at |
AT5G19875
|
unknown |
-0.129 |
-0.107 |
| 79 |
250205_at |
AT5G14020
|
[Endosomal targeting BRO1-like domain-containing protein] |
-0.127 |
0.051 |
| 80 |
255198_at |
AT4G07460
|
unknown |
-0.126 |
0.006 |
| 81 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.125 |
-0.082 |
| 82 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.124 |
-0.021 |
| 83 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.124 |
-0.117 |
| 84 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.123 |
0.005 |
| 85 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.123 |
0.043 |
| 86 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.122 |
0.125 |
| 87 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
-0.122 |
-0.113 |
| 88 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.122 |
-0.048 |
| 89 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.122 |
0.049 |