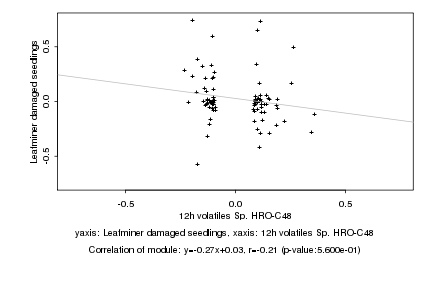
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
12h volatiles Sp. HRO-C48 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
261658_at |
AT1G50040
|
unknown |
0.359 |
-0.112 |
| 2 |
254043_at |
AT4G25990
|
CIL |
0.343 |
-0.282 |
| 3 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.263 |
0.496 |
| 4 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.251 |
0.168 |
| 5 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.220 |
-0.181 |
| 6 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
0.191 |
0.019 |
| 7 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.191 |
-0.062 |
| 8 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
0.186 |
-0.211 |
| 9 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.184 |
-0.029 |
| 10 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
0.153 |
-0.284 |
| 11 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.153 |
0.026 |
| 12 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
0.150 |
0.031 |
| 13 |
260939_at |
AT1G45180
|
[RING/U-box superfamily protein] |
0.140 |
-0.021 |
| 14 |
267267_at |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
0.139 |
0.061 |
| 15 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
0.131 |
-0.026 |
| 16 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
0.129 |
-0.092 |
| 17 |
254529_at |
AT4G19540
|
INDH, iron-sulfur protein required for NADH dehydrogenase, INDL, IND1(iron-sulfur protein required for NADH dehydrogenase)-like |
0.121 |
-0.170 |
| 18 |
252217_at |
AT3G50140
|
unknown |
0.118 |
0.005 |
| 19 |
252432_at |
AT3G47680
|
[DNA binding] |
0.118 |
-0.023 |
| 20 |
262213_at |
AT1G74870
|
[RING/U-box superfamily protein] |
0.117 |
-0.100 |
| 21 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.117 |
-0.050 |
| 22 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
0.113 |
-0.288 |
| 23 |
249445_at |
AT5G39380
|
[Plant calmodulin-binding protein-related] |
0.112 |
0.024 |
| 24 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.110 |
0.732 |
| 25 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.110 |
0.058 |
| 26 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
0.108 |
-0.413 |
| 27 |
257139_at |
AT3G28890
|
AtRLP43, receptor like protein 43, RLP43, receptor like protein 43 |
0.107 |
0.167 |
| 28 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
0.106 |
0.005 |
| 29 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
0.103 |
0.034 |
| 30 |
263362_at |
AT2G03880
|
REME1, required for efficiency of mitochondrial editing 1 |
0.100 |
-0.000 |
| 31 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.100 |
-0.066 |
| 32 |
256262_at |
AT3G12150
|
unknown |
0.100 |
-0.250 |
| 33 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.099 |
0.650 |
| 34 |
245813_at |
AT1G49920
|
[MuDR family transposase] |
0.098 |
-0.008 |
| 35 |
263387_at |
AT2G40160
|
TBL30 |
0.096 |
0.026 |
| 36 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.092 |
0.339 |
| 37 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.091 |
0.003 |
| 38 |
259615_at |
AT1G47980
|
unknown |
0.091 |
0.012 |
| 39 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.090 |
0.015 |
| 40 |
264879_at |
AT1G61260
|
unknown |
0.089 |
0.047 |
| 41 |
253583_at |
AT4G30680
|
[Initiation factor eIF-4 gamma, MA3] |
0.089 |
0.003 |
| 42 |
261856_at |
AT1G50530
|
unknown |
0.087 |
0.020 |
| 43 |
248441_at |
AT5G51270
|
[U-box domain-containing protein kinase family protein] |
0.087 |
-0.026 |
| 44 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.086 |
-0.083 |
| 45 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
0.085 |
-0.178 |
| 46 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
0.084 |
-0.035 |
| 47 |
257352_at |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
0.083 |
-0.010 |
| 48 |
257014_at |
AT3G26930
|
[Protein with RNI-like/FBD-like domains] |
0.081 |
-0.064 |
| 49 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.232 |
0.284 |
| 50 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.215 |
-0.008 |
| 51 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.198 |
0.749 |
| 52 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
-0.196 |
0.236 |
| 53 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.179 |
0.090 |
| 54 |
265892_at |
AT2G15020
|
unknown |
-0.175 |
-0.571 |
| 55 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.174 |
0.384 |
| 56 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
-0.152 |
0.326 |
| 57 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.147 |
0.007 |
| 58 |
253807_at |
AT4G28280
|
LLG3, LORELEI-LIKE-GPI ANCHORED PROTEIN 3 |
-0.142 |
0.126 |
| 59 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.140 |
0.213 |
| 60 |
255873_at |
AT2G30340
|
LBD13, LOB domain-containing protein 13 |
-0.139 |
-0.029 |
| 61 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.134 |
0.097 |
| 62 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.134 |
-0.025 |
| 63 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
-0.133 |
-0.019 |
| 64 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.131 |
-0.316 |
| 65 |
246739_at |
AT5G27800
|
[Class II aminoacyl-tRNA and biotin synthetases superfamily protein] |
-0.131 |
-0.011 |
| 66 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
-0.130 |
0.019 |
| 67 |
263642_at |
AT2G04750
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.128 |
0.013 |
| 68 |
256059_at |
AT1G06990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.121 |
-0.047 |
| 69 |
257869_at |
AT3G25160
|
[ER lumen protein retaining receptor family protein] |
-0.119 |
0.015 |
| 70 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.118 |
-0.206 |
| 71 |
265590_at |
AT2G20160
|
ASK17, ARABIDOPSIS SKP1-LIKE 17, MEO, MEIDOS |
-0.117 |
-0.008 |
| 72 |
264068_at |
AT2G27990
|
BLH8, BEL1-like homeodomain 8, PNF, POUND-FOOLISH |
-0.115 |
-0.161 |
| 73 |
257693_at |
AT3G12850
|
[COP9 signalosome complex-related / CSN complex-related] |
-0.115 |
-0.008 |
| 74 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.111 |
0.336 |
| 75 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.111 |
-0.008 |
| 76 |
255014_at |
AT4G09960
|
AGL11, AGAMOUS-like 11, STK, SEEDSTICK |
-0.111 |
-0.007 |
| 77 |
257032_at |
AT3G19140
|
DNF, DAY NEUTRAL FLOWERING |
-0.107 |
-0.020 |
| 78 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.107 |
0.216 |
| 79 |
252817_at |
AT3G42580
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G12100.1)] |
-0.106 |
-0.056 |
| 80 |
251825_at |
AT3G55100
|
ABCG17, ATP-binding cassette G17 |
-0.106 |
-0.009 |
| 81 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
-0.105 |
0.598 |
| 82 |
252538_at |
AT3G45720
|
[Major facilitator superfamily protein] |
-0.105 |
0.001 |
| 83 |
257531_at |
AT3G07250
|
[nuclear transport factor 2 (NTF2) family protein / RNA recognition motif (RRM)-containing protein] |
-0.104 |
0.011 |
| 84 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
-0.103 |
0.040 |
| 85 |
266755_at |
AT2G47150
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.103 |
-0.004 |
| 86 |
247599_at |
AT5G60880
|
BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE |
-0.103 |
-0.022 |
| 87 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.102 |
0.110 |
| 88 |
257331_at |
ATMG01330
|
ORF107H |
-0.102 |
-0.021 |
| 89 |
267146_at |
AT2G38160
|
unknown |
-0.102 |
-0.073 |
| 90 |
247230_at |
AT5G65170
|
[VQ motif-containing protein] |
-0.101 |
0.044 |
| 91 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.101 |
0.225 |
| 92 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.099 |
0.267 |
| 93 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
-0.096 |
0.018 |
| 94 |
259926_at |
AT1G75090
|
[DNA glycosylase superfamily protein] |
-0.095 |
-0.048 |
| 95 |
245669_at |
AT1G28300
|
LEC2, LEAFY COTYLEDON 2 |
-0.094 |
-0.075 |