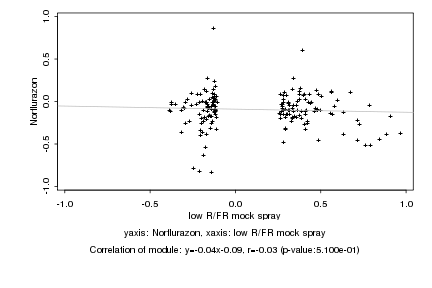
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
low R/FR mock spray |
Log2 signal ratio
Norflurazon |
| 1 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.965 |
-0.369 |
| 2 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.905 |
-0.168 |
| 3 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.885 |
-0.388 |
| 4 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.842 |
-0.439 |
| 5 |
253155_at |
AT4G35720
|
unknown |
0.789 |
-0.512 |
| 6 |
251012_at |
AT5G02580
|
unknown |
0.785 |
-0.038 |
| 7 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.759 |
-0.510 |
| 8 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.724 |
-0.268 |
| 9 |
247878_at |
AT5G57760
|
unknown |
0.715 |
-0.452 |
| 10 |
250327_at |
AT5G12050
|
unknown |
0.712 |
-0.214 |
| 11 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.669 |
0.112 |
| 12 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.632 |
-0.382 |
| 13 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.629 |
-0.126 |
| 14 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.595 |
0.018 |
| 15 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.578 |
-0.058 |
| 16 |
255413_at |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.564 |
-0.146 |
| 17 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.562 |
0.117 |
| 18 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.558 |
0.120 |
| 19 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.555 |
-0.140 |
| 20 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.503 |
0.067 |
| 21 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.494 |
-0.105 |
| 22 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.485 |
0.085 |
| 23 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.482 |
-0.451 |
| 24 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.478 |
-0.091 |
| 25 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.476 |
-0.097 |
| 26 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.475 |
0.140 |
| 27 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.466 |
-0.074 |
| 28 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.461 |
-0.110 |
| 29 |
261456_at |
AT1G21050
|
unknown |
0.443 |
-0.003 |
| 30 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.437 |
-0.023 |
| 31 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.431 |
0.093 |
| 32 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.426 |
-0.003 |
| 33 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.418 |
-0.250 |
| 34 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.417 |
-0.226 |
| 35 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
0.411 |
-0.101 |
| 36 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.409 |
-0.149 |
| 37 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
0.408 |
0.024 |
| 38 |
265276_at |
AT2G28400
|
unknown |
0.406 |
-0.326 |
| 39 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.405 |
-0.107 |
| 40 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.403 |
-0.263 |
| 41 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.400 |
0.087 |
| 42 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.397 |
0.082 |
| 43 |
245076_at |
AT2G23170
|
GH3.3 |
0.391 |
0.611 |
| 44 |
260883_at |
AT1G29270
|
unknown |
0.387 |
-0.192 |
| 45 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.386 |
-0.107 |
| 46 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.377 |
0.158 |
| 47 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.375 |
0.031 |
| 48 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.372 |
-0.160 |
| 49 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.371 |
0.085 |
| 50 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.370 |
0.121 |
| 51 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.363 |
-0.100 |
| 52 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.359 |
0.003 |
| 53 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.357 |
-0.044 |
| 54 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.354 |
-0.178 |
| 55 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.343 |
-0.168 |
| 56 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.338 |
0.273 |
| 57 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.336 |
-0.036 |
| 58 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.335 |
-0.109 |
| 59 |
252204_at |
AT3G50340
|
unknown |
0.333 |
-0.180 |
| 60 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.332 |
-0.071 |
| 61 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.331 |
0.151 |
| 62 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.331 |
-0.196 |
| 63 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.324 |
-0.226 |
| 64 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.316 |
-0.149 |
| 65 |
258367_at |
AT3G14370
|
WAG2 |
0.313 |
-0.096 |
| 66 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.312 |
-0.037 |
| 67 |
245439_at |
AT4G16670
|
unknown |
0.308 |
-0.001 |
| 68 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.306 |
-0.020 |
| 69 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.306 |
-0.097 |
| 70 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.304 |
-0.148 |
| 71 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.298 |
0.080 |
| 72 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.295 |
-0.143 |
| 73 |
248509_at |
AT5G50335
|
unknown |
0.292 |
-0.307 |
| 74 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.290 |
-0.084 |
| 75 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.289 |
-0.182 |
| 76 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.288 |
-0.324 |
| 77 |
256114_at |
AT1G16850
|
unknown |
0.287 |
0.111 |
| 78 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.281 |
0.077 |
| 79 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.280 |
-0.006 |
| 80 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.279 |
-0.143 |
| 81 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.279 |
0.028 |
| 82 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
0.278 |
-0.096 |
| 83 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.278 |
-0.472 |
| 84 |
263169_at |
AT1G03010
|
[Phototropic-responsive NPH3 family protein] |
0.274 |
-0.047 |
| 85 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.271 |
-0.079 |
| 86 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.270 |
-0.031 |
| 87 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.268 |
-0.106 |
| 88 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.266 |
-0.034 |
| 89 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.263 |
-0.196 |
| 90 |
251010_at |
AT5G02550
|
unknown |
0.261 |
-0.144 |
| 91 |
263151_at |
AT1G54120
|
unknown |
0.261 |
0.093 |
| 92 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.258 |
-0.130 |
| 93 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.392 |
-0.101 |
| 94 |
260081_at |
AT1G78170
|
unknown |
-0.382 |
-0.117 |
| 95 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.377 |
-0.008 |
| 96 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.376 |
-0.032 |
| 97 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.356 |
-0.029 |
| 98 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.321 |
-0.353 |
| 99 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.317 |
-0.101 |
| 100 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.308 |
-0.070 |
| 101 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.304 |
-0.073 |
| 102 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.298 |
-0.008 |
| 103 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.295 |
-0.250 |
| 104 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.284 |
0.032 |
| 105 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.272 |
-0.226 |
| 106 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.261 |
0.097 |
| 107 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.260 |
-0.039 |
| 108 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.251 |
-0.786 |
| 109 |
262170_at |
AT1G74940
|
unknown |
-0.230 |
-0.031 |
| 110 |
245341_at |
AT4G16447
|
unknown |
-0.225 |
0.085 |
| 111 |
248917_at |
AT5G45850
|
unknown |
-0.215 |
-0.006 |
| 112 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.213 |
-0.818 |
| 113 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.211 |
-0.147 |
| 114 |
254145_at |
AT4G24700
|
unknown |
-0.210 |
-0.390 |
| 115 |
256096_at |
AT1G13650
|
unknown |
-0.210 |
-0.339 |
| 116 |
252057_at |
AT3G52480
|
unknown |
-0.206 |
0.083 |
| 117 |
262811_at |
AT1G11700
|
unknown |
-0.199 |
-0.255 |
| 118 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.199 |
-0.193 |
| 119 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.197 |
0.002 |
| 120 |
245776_at |
AT1G30260
|
unknown |
-0.195 |
-0.364 |
| 121 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.192 |
-0.634 |
| 122 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.190 |
-0.234 |
| 123 |
246913_at |
AT5G25830
|
GATA12, GATA transcription factor 12 |
-0.188 |
-0.095 |
| 124 |
262162_at |
AT1G78020
|
unknown |
-0.186 |
-0.186 |
| 125 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.186 |
0.142 |
| 126 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
-0.177 |
0.000 |
| 127 |
264096_at |
AT1G78995
|
unknown |
-0.176 |
-0.531 |
| 128 |
262421_at |
AT1G50290
|
unknown |
-0.175 |
-0.009 |
| 129 |
259179_at |
AT3G01660
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.175 |
-0.380 |
| 130 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.174 |
-0.005 |
| 131 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
-0.172 |
0.124 |
| 132 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.170 |
-0.200 |
| 133 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.168 |
-0.108 |
| 134 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
-0.168 |
-0.032 |
| 135 |
259887_at |
AT1G76360
|
[Protein kinase superfamily protein] |
-0.168 |
-0.068 |
| 136 |
246781_at |
AT5G27350
|
SFP1 |
-0.167 |
-0.048 |
| 137 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.167 |
0.276 |
| 138 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.163 |
-0.080 |
| 139 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.161 |
-0.175 |
| 140 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.161 |
-0.052 |
| 141 |
253836_at |
AT4G27840
|
[SNARE-like superfamily protein] |
-0.161 |
-0.066 |
| 142 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.155 |
-0.161 |
| 143 |
248032_at |
AT5G55860
|
unknown |
-0.154 |
0.026 |
| 144 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.149 |
-0.309 |
| 145 |
262598_at |
AT1G15260
|
unknown |
-0.147 |
-0.173 |
| 146 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.146 |
-0.047 |
| 147 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.144 |
-0.832 |
| 148 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.143 |
-0.254 |
| 149 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-0.141 |
-0.169 |
| 150 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.141 |
-0.086 |
| 151 |
252569_at |
AT3G45420
|
LecRK-I.4, L-type lectin receptor kinase I.4 |
-0.140 |
-0.009 |
| 152 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.139 |
0.102 |
| 153 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.138 |
-0.231 |
| 154 |
253326_at |
AT4G33440
|
[Pectin lyase-like superfamily protein] |
-0.138 |
0.051 |
| 155 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.134 |
-0.045 |
| 156 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.134 |
0.862 |
| 157 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
-0.132 |
-0.007 |
| 158 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.132 |
-0.025 |
| 159 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.130 |
0.040 |
| 160 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.130 |
0.149 |
| 161 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.129 |
-0.045 |
| 162 |
252661_at |
AT3G44450
|
unknown |
-0.129 |
0.044 |
| 163 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
-0.126 |
-0.087 |
| 164 |
253038_at |
AT4G37790
|
HAT22 |
-0.126 |
-0.117 |
| 165 |
265664_at |
AT2G24420
|
[DNA repair ATPase-related] |
-0.126 |
0.037 |
| 166 |
263135_at |
AT1G78550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.125 |
0.066 |
| 167 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.124 |
0.011 |
| 168 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.124 |
-0.121 |
| 169 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.124 |
0.023 |
| 170 |
255171_at |
AT4G07990
|
[Chaperone DnaJ-domain superfamily protein] |
-0.123 |
0.240 |
| 171 |
246217_at |
AT4G36920
|
AP2, APETALA 2, AtAP2, FL1, FLOWER 1, FLO2, FLORAL MUTANT 2 |
-0.123 |
0.083 |
| 172 |
262650_at |
AT1G14090
|
[pseudogene] |
-0.120 |
0.180 |
| 173 |
249006_at |
AT5G44660
|
unknown |
-0.119 |
-0.055 |
| 174 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
-0.118 |
-0.092 |
| 175 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.118 |
0.035 |
| 176 |
261120_at |
AT1G75410
|
BLH3, BEL1-like homeodomain 3 |
-0.118 |
-0.108 |
| 177 |
262711_at |
AT1G16500
|
unknown |
-0.117 |
-0.099 |
| 178 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.117 |
-0.104 |
| 179 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.117 |
-0.142 |
| 180 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
-0.116 |
0.069 |
| 181 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.116 |
-0.328 |
| 182 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.113 |
-0.178 |
| 183 |
250524_at |
AT5G08520
|
[Duplicated homeodomain-like superfamily protein] |
-0.111 |
-0.010 |