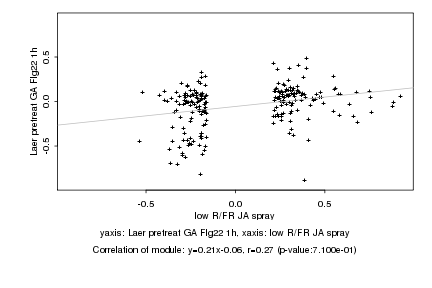
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
low R/FR JA spray |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.921 |
0.066 |
| 2 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.883 |
-0.008 |
| 3 |
251012_at |
AT5G02580
|
unknown |
0.876 |
-0.051 |
| 4 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.757 |
-0.119 |
| 5 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.750 |
0.049 |
| 6 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.746 |
0.115 |
| 7 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.678 |
-0.234 |
| 8 |
253155_at |
AT4G35720
|
unknown |
0.675 |
0.106 |
| 9 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.659 |
-0.164 |
| 10 |
247878_at |
AT5G57760
|
unknown |
0.634 |
-0.023 |
| 11 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.586 |
0.089 |
| 12 |
250327_at |
AT5G12050
|
unknown |
0.579 |
-0.147 |
| 13 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.574 |
0.090 |
| 14 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.556 |
0.147 |
| 15 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.551 |
0.137 |
| 16 |
255413_at |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.544 |
-0.106 |
| 17 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.543 |
0.291 |
| 18 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.487 |
-0.022 |
| 19 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.481 |
0.050 |
| 20 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.475 |
0.106 |
| 21 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
0.467 |
0.047 |
| 22 |
256442_at |
AT3G10930
|
unknown |
0.451 |
0.087 |
| 23 |
265276_at |
AT2G28400
|
unknown |
0.444 |
0.023 |
| 24 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.434 |
0.013 |
| 25 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.427 |
0.044 |
| 26 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.419 |
-0.038 |
| 27 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.407 |
-0.199 |
| 28 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.404 |
-0.432 |
| 29 |
245076_at |
AT2G23170
|
GH3.3 |
0.397 |
0.484 |
| 30 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.393 |
0.372 |
| 31 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.390 |
0.047 |
| 32 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.383 |
-0.878 |
| 33 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.383 |
0.090 |
| 34 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.376 |
0.279 |
| 35 |
260883_at |
AT1G29270
|
unknown |
0.371 |
0.097 |
| 36 |
245439_at |
AT4G16670
|
unknown |
0.367 |
0.077 |
| 37 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.364 |
0.014 |
| 38 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.354 |
0.108 |
| 39 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.349 |
0.118 |
| 40 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.348 |
0.413 |
| 41 |
261456_at |
AT1G21050
|
unknown |
0.347 |
-0.091 |
| 42 |
255642_at |
AT4G00820
|
iqd17, IQ-domain 17 |
0.343 |
0.127 |
| 43 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.342 |
0.179 |
| 44 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.339 |
0.101 |
| 45 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.333 |
0.020 |
| 46 |
249885_at |
AT5G22940
|
F8H, FRA8 homolog |
0.327 |
0.064 |
| 47 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.326 |
0.091 |
| 48 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.324 |
0.155 |
| 49 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.322 |
-0.372 |
| 50 |
249887_at |
AT5G22310
|
unknown |
0.320 |
0.084 |
| 51 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.316 |
0.130 |
| 52 |
252204_at |
AT3G50340
|
unknown |
0.316 |
-0.018 |
| 53 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.312 |
-0.043 |
| 54 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
0.310 |
-0.307 |
| 55 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.310 |
-0.101 |
| 56 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.310 |
0.098 |
| 57 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.307 |
0.135 |
| 58 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.305 |
-0.224 |
| 59 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.305 |
-0.131 |
| 60 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
0.303 |
0.166 |
| 61 |
263169_at |
AT1G03010
|
[Phototropic-responsive NPH3 family protein] |
0.302 |
-0.005 |
| 62 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.301 |
0.382 |
| 63 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.300 |
-0.354 |
| 64 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.299 |
0.075 |
| 65 |
261292_at |
AT1G36940
|
unknown |
0.296 |
0.073 |
| 66 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
0.294 |
0.060 |
| 67 |
248003_at |
AT5G56220
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.294 |
0.125 |
| 68 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.294 |
0.239 |
| 69 |
247743_at |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
0.288 |
0.078 |
| 70 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.285 |
0.070 |
| 71 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.285 |
-0.031 |
| 72 |
258702_at |
AT3G09730
|
unknown |
0.281 |
0.144 |
| 73 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.276 |
0.114 |
| 74 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
0.274 |
0.054 |
| 75 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.270 |
0.180 |
| 76 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.265 |
-0.131 |
| 77 |
260470_at |
AT1G11125
|
unknown |
0.264 |
-0.004 |
| 78 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.264 |
0.201 |
| 79 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.264 |
0.016 |
| 80 |
247349_at |
AT5G63820
|
unknown |
0.263 |
0.104 |
| 81 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
0.260 |
0.087 |
| 82 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
0.260 |
0.066 |
| 83 |
264264_at |
AT1G09250
|
AIF4, ATBS1 Interacting Factor 4 |
0.256 |
-0.044 |
| 84 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
0.256 |
-0.161 |
| 85 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.254 |
0.117 |
| 86 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
0.253 |
-0.211 |
| 87 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.253 |
-0.143 |
| 88 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.251 |
0.111 |
| 89 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.249 |
0.009 |
| 90 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.245 |
0.065 |
| 91 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.245 |
0.104 |
| 92 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.242 |
0.044 |
| 93 |
263151_at |
AT1G54120
|
unknown |
0.239 |
-0.168 |
| 94 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.239 |
0.205 |
| 95 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.238 |
0.034 |
| 96 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.237 |
0.114 |
| 97 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.233 |
0.044 |
| 98 |
259823_at |
AT1G66250
|
[O-Glycosyl hydrolases family 17 protein] |
0.231 |
-0.142 |
| 99 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.231 |
0.363 |
| 100 |
247596_at |
AT5G60840
|
unknown |
0.225 |
-0.066 |
| 101 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.225 |
0.147 |
| 102 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.222 |
-0.166 |
| 103 |
251643_at |
AT3G57550
|
AGK2, guanylate kinase, GK-2, GUANYLATE KINAS 2 |
0.220 |
0.145 |
| 104 |
252133_at |
AT3G50900
|
unknown |
0.220 |
0.122 |
| 105 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
0.219 |
0.047 |
| 106 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
0.218 |
0.119 |
| 107 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.217 |
-0.094 |
| 108 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
0.216 |
0.014 |
| 109 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.211 |
-0.166 |
| 110 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.210 |
-0.240 |
| 111 |
251010_at |
AT5G02550
|
unknown |
0.210 |
0.437 |
| 112 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.539 |
-0.443 |
| 113 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.524 |
0.106 |
| 114 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
-0.428 |
0.068 |
| 115 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.401 |
0.120 |
| 116 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.401 |
0.018 |
| 117 |
245341_at |
AT4G16447
|
unknown |
-0.384 |
0.003 |
| 118 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.374 |
-0.538 |
| 119 |
263674_at |
AT2G04790
|
unknown |
-0.366 |
-0.694 |
| 120 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.363 |
0.034 |
| 121 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.357 |
-0.281 |
| 122 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.353 |
-0.442 |
| 123 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.344 |
-0.113 |
| 124 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.331 |
0.003 |
| 125 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.331 |
0.110 |
| 126 |
256891_at |
AT3G19030
|
unknown |
-0.330 |
-0.100 |
| 127 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.328 |
-0.698 |
| 128 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.315 |
-0.028 |
| 129 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.315 |
0.064 |
| 130 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.314 |
-0.507 |
| 131 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.309 |
-0.176 |
| 132 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.306 |
0.212 |
| 133 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.300 |
-0.597 |
| 134 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.299 |
-0.574 |
| 135 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.296 |
-0.030 |
| 136 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.293 |
-0.428 |
| 137 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.293 |
-0.296 |
| 138 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.290 |
0.021 |
| 139 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.289 |
0.068 |
| 140 |
262162_at |
AT1G78020
|
unknown |
-0.286 |
-0.350 |
| 141 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.284 |
-0.625 |
| 142 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.282 |
-0.017 |
| 143 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.281 |
0.101 |
| 144 |
260081_at |
AT1G78170
|
unknown |
-0.281 |
0.054 |
| 145 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.279 |
0.011 |
| 146 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.273 |
-0.431 |
| 147 |
259520_at |
AT1G12320
|
unknown |
-0.271 |
0.175 |
| 148 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.271 |
-0.010 |
| 149 |
252938_at |
AT4G39190
|
unknown |
-0.270 |
0.186 |
| 150 |
266456_at |
AT2G22770
|
NAI1 |
-0.269 |
0.072 |
| 151 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.265 |
-0.486 |
| 152 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
-0.265 |
-0.004 |
| 153 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.259 |
-0.466 |
| 154 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.256 |
-0.406 |
| 155 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
-0.255 |
0.062 |
| 156 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.254 |
-0.216 |
| 157 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.253 |
0.124 |
| 158 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.252 |
-0.040 |
| 159 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.250 |
-0.181 |
| 160 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.249 |
-0.472 |
| 161 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.248 |
0.001 |
| 162 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.244 |
-0.005 |
| 163 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.241 |
-0.115 |
| 164 |
265545_at |
AT2G28250
|
NCRK |
-0.241 |
0.083 |
| 165 |
247486_at |
AT5G62140
|
unknown |
-0.239 |
-0.441 |
| 166 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.238 |
0.077 |
| 167 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.234 |
-0.025 |
| 168 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.233 |
0.124 |
| 169 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.223 |
0.108 |
| 170 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
-0.223 |
0.070 |
| 171 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.220 |
-0.065 |
| 172 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.218 |
0.099 |
| 173 |
253038_at |
AT4G37790
|
HAT22 |
-0.218 |
0.047 |
| 174 |
252619_at |
AT3G45210
|
unknown |
-0.215 |
-0.001 |
| 175 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.214 |
0.044 |
| 176 |
252097_at |
AT3G51090
|
unknown |
-0.211 |
0.230 |
| 177 |
262392_at |
AT1G49520
|
[SWIB complex BAF60b domain-containing protein] |
-0.209 |
0.011 |
| 178 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.206 |
-0.049 |
| 179 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.204 |
-0.488 |
| 180 |
245776_at |
AT1G30260
|
unknown |
-0.202 |
-0.112 |
| 181 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.202 |
-0.087 |
| 182 |
247605_at |
AT5G60970
|
TCP5, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 |
-0.201 |
0.013 |
| 183 |
248020_at |
AT5G56490
|
AtGulLO4, GulLO4, L -gulono-1,4-lactone ( L -GulL) oxidase 4 |
-0.200 |
0.080 |
| 184 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.197 |
-0.811 |
| 185 |
251194_at |
AT3G62920
|
unknown |
-0.196 |
0.209 |
| 186 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.196 |
-0.392 |
| 187 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.195 |
-0.354 |
| 188 |
259207_at |
AT3G09050
|
unknown |
-0.195 |
-0.412 |
| 189 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.193 |
-0.354 |
| 190 |
250908_at |
AT5G03680
|
PTL, PETAL LOSS |
-0.193 |
0.061 |
| 191 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.192 |
0.039 |
| 192 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
-0.191 |
0.083 |
| 193 |
262598_at |
AT1G15260
|
unknown |
-0.191 |
-0.386 |
| 194 |
260139_at |
AT1G66380
|
AtMYB114, myb domain protein 114, MYB114, myb domain protein 114 |
-0.191 |
0.072 |
| 195 |
253836_at |
AT4G27840
|
[SNARE-like superfamily protein] |
-0.191 |
0.329 |
| 196 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
-0.190 |
0.277 |
| 197 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.190 |
0.025 |
| 198 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
-0.190 |
-0.138 |
| 199 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.189 |
-0.593 |
| 200 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.187 |
-0.111 |
| 201 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.185 |
0.094 |
| 202 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.183 |
-0.261 |
| 203 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.178 |
-0.118 |
| 204 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.177 |
-0.544 |
| 205 |
246948_at |
AT5G25130
|
CYP71B12, cytochrome P450, family 71, subfamily B, polypeptide 12 |
-0.176 |
-0.092 |
| 206 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
-0.176 |
-0.023 |
| 207 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.175 |
-0.086 |
| 208 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.174 |
0.037 |
| 209 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
-0.173 |
0.059 |
| 210 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.173 |
-0.001 |
| 211 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.172 |
-0.494 |
| 212 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.171 |
0.186 |
| 213 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.171 |
-0.250 |
| 214 |
264385_at |
AT1G12020
|
unknown |
-0.170 |
-0.114 |
| 215 |
262170_at |
AT1G74940
|
unknown |
-0.170 |
-0.056 |
| 216 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
-0.168 |
0.289 |
| 217 |
249134_at |
AT5G43150
|
unknown |
-0.167 |
-0.398 |
| 218 |
260159_at |
AT1G79890
|
[RAD3-like DNA-binding helicase protein] |
-0.167 |
-0.001 |
| 219 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.167 |
-0.134 |
| 220 |
259528_at |
AT1G12330
|
unknown |
-0.167 |
-0.206 |
| 221 |
249946_at |
AT5G19170
|
unknown |
-0.166 |
0.077 |