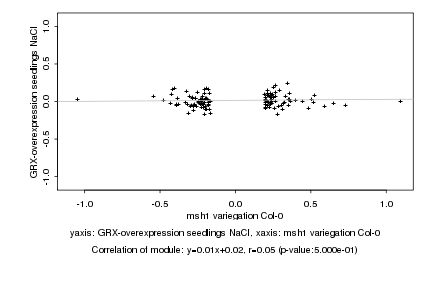
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msh1 variegation Col-0 |
Log2 signal ratio
GRX-overexpression seedlings NaCl |
| 1 |
262719_at |
AT1G43590
|
unknown |
1.092 |
0.005 |
| 2 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.729 |
-0.047 |
| 3 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.647 |
-0.021 |
| 4 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.589 |
-0.058 |
| 5 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.522 |
0.081 |
| 6 |
260522_x_at |
AT2G41730
|
unknown |
0.512 |
-0.006 |
| 7 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.503 |
0.035 |
| 8 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.478 |
-0.091 |
| 9 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.444 |
0.007 |
| 10 |
263515_at |
AT2G21640
|
unknown |
0.396 |
0.023 |
| 11 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.365 |
0.007 |
| 12 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.354 |
0.027 |
| 13 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.353 |
0.111 |
| 14 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.350 |
0.028 |
| 15 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.349 |
-0.043 |
| 16 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.341 |
0.247 |
| 17 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.326 |
0.072 |
| 18 |
244906_at |
ATMG00690
|
ORF240A |
0.321 |
-0.012 |
| 19 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.317 |
-0.025 |
| 20 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.308 |
-0.100 |
| 21 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.300 |
-0.044 |
| 22 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.292 |
0.160 |
| 23 |
251595_at |
AT3G57620
|
[glyoxal oxidase-related protein] |
0.282 |
-0.057 |
| 24 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.273 |
-0.164 |
| 25 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.265 |
0.225 |
| 26 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.264 |
0.080 |
| 27 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.262 |
0.013 |
| 28 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.260 |
0.121 |
| 29 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.258 |
-0.087 |
| 30 |
244901_at |
ATMG00640
|
ORF25 |
0.249 |
0.057 |
| 31 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.248 |
0.065 |
| 32 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.247 |
0.197 |
| 33 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.241 |
0.101 |
| 34 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.239 |
-0.016 |
| 35 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.237 |
0.012 |
| 36 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.236 |
0.034 |
| 37 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.233 |
-0.015 |
| 38 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.232 |
-0.038 |
| 39 |
256420_at |
AT1G33475
|
[SNARE-like superfamily protein] |
0.230 |
0.067 |
| 40 |
263853_at |
AT2G04440
|
[MutT/nudix family protein] |
0.229 |
0.082 |
| 41 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.228 |
0.115 |
| 42 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
0.227 |
-0.002 |
| 43 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
0.227 |
-0.013 |
| 44 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.226 |
0.070 |
| 45 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.226 |
0.064 |
| 46 |
266658_at |
AT2G25735
|
unknown |
0.225 |
-0.069 |
| 47 |
249634_at |
AT5G36840
|
unknown |
0.222 |
0.073 |
| 48 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
0.217 |
-0.011 |
| 49 |
253265_at |
AT4G34040
|
MBR2, MED25 BINDING RING-H2 PROTEIN 2 |
0.215 |
-0.008 |
| 50 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.213 |
0.084 |
| 51 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.210 |
0.153 |
| 52 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.209 |
-0.042 |
| 53 |
244921_s_at |
ATMG01000
|
ORF114 |
0.209 |
0.116 |
| 54 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.207 |
0.083 |
| 55 |
249889_at |
AT5G22540
|
unknown |
0.205 |
0.013 |
| 56 |
247670_at |
AT5G60190
|
[Cysteine proteinases superfamily protein] |
0.205 |
0.017 |
| 57 |
257334_at |
ATMG01370
|
ORF111D |
0.205 |
0.014 |
| 58 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.199 |
-0.079 |
| 59 |
265169_x_at |
AT1G23720
|
[Proline-rich extensin-like family protein] |
0.199 |
0.064 |
| 60 |
247958_at |
AT5G57070
|
[hydroxyproline-rich glycoprotein family protein] |
0.199 |
0.022 |
| 61 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
0.197 |
-0.008 |
| 62 |
249981_at |
AT5G18510
|
[Aminotransferase-like, plant mobile domain family protein] |
0.196 |
0.037 |
| 63 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
0.195 |
-0.083 |
| 64 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.195 |
-0.036 |
| 65 |
261972_at |
AT1G64600
|
[methyltransferases] |
0.192 |
0.103 |
| 66 |
267459_at |
AT2G33850
|
unknown |
-1.054 |
0.037 |
| 67 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.545 |
0.080 |
| 68 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.482 |
0.015 |
| 69 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.432 |
-0.025 |
| 70 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.427 |
0.096 |
| 71 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.422 |
0.161 |
| 72 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.410 |
0.181 |
| 73 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.396 |
-0.030 |
| 74 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.392 |
-0.040 |
| 75 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.389 |
0.046 |
| 76 |
258731_at |
AT3G11880
|
unknown |
-0.380 |
-0.039 |
| 77 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
-0.332 |
-0.010 |
| 78 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.326 |
0.136 |
| 79 |
255527_at |
AT4G02360
|
unknown |
-0.322 |
-0.035 |
| 80 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
-0.312 |
-0.148 |
| 81 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.308 |
0.072 |
| 82 |
251646_at |
AT3G57780
|
unknown |
-0.304 |
-0.055 |
| 83 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.298 |
-0.049 |
| 84 |
249378_at |
AT5G40450
|
unknown |
-0.291 |
0.044 |
| 85 |
254724_at |
AT4G13630
|
unknown |
-0.290 |
0.066 |
| 86 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.284 |
-0.056 |
| 87 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.283 |
-0.043 |
| 88 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.283 |
-0.119 |
| 89 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-0.275 |
-0.034 |
| 90 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
-0.274 |
-0.052 |
| 91 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.270 |
0.052 |
| 92 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.269 |
0.045 |
| 93 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.259 |
-0.061 |
| 94 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.257 |
0.124 |
| 95 |
248032_at |
AT5G55860
|
unknown |
-0.246 |
-0.007 |
| 96 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.246 |
0.003 |
| 97 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
-0.240 |
-0.023 |
| 98 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.233 |
-0.036 |
| 99 |
250982_at |
AT5G03150
|
JKD, JACKDAW |
-0.232 |
0.029 |
| 100 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.230 |
-0.014 |
| 101 |
246759_at |
AT5G27950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.229 |
-0.070 |
| 102 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.227 |
0.035 |
| 103 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.226 |
0.002 |
| 104 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.223 |
0.036 |
| 105 |
255876_at |
AT2G40480
|
unknown |
-0.223 |
-0.031 |
| 106 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.222 |
0.078 |
| 107 |
250696_at |
AT5G06790
|
unknown |
-0.217 |
-0.079 |
| 108 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.215 |
-0.034 |
| 109 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.211 |
0.116 |
| 110 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.211 |
0.025 |
| 111 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.209 |
0.164 |
| 112 |
249167_at |
AT5G42860
|
unknown |
-0.208 |
-0.009 |
| 113 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.207 |
-0.163 |
| 114 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.203 |
0.047 |
| 115 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.200 |
-0.103 |
| 116 |
261151_at |
AT1G19650
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.200 |
-0.065 |
| 117 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.197 |
-0.105 |
| 118 |
266456_at |
AT2G22770
|
NAI1 |
-0.192 |
0.174 |
| 119 |
263972_at |
AT2G42760
|
unknown |
-0.192 |
0.042 |
| 120 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.189 |
0.029 |
| 121 |
245353_at |
AT4G16000
|
unknown |
-0.185 |
-0.044 |
| 122 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
-0.184 |
-0.036 |
| 123 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.179 |
-0.002 |
| 124 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.179 |
0.167 |
| 125 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.175 |
0.002 |
| 126 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.175 |
0.107 |
| 127 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.174 |
-0.093 |
| 128 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.171 |
-0.154 |
| 129 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.170 |
0.009 |