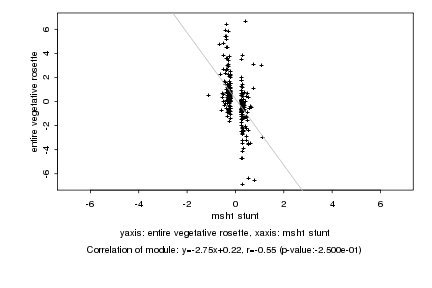
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
msh1 stunt |
Log2 signal ratio
entire vegetative rosette |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.089 |
-2.937 |
| 2 |
262719_at |
AT1G43590
|
unknown |
1.074 |
3.053 |
| 3 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.758 |
-6.505 |
| 4 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.747 |
3.157 |
| 5 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.716 |
1.156 |
| 6 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.661 |
-0.432 |
| 7 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.603 |
-3.469 |
| 8 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.585 |
-0.395 |
| 9 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.575 |
-0.571 |
| 10 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.569 |
-0.556 |
| 11 |
263475_at |
AT2G31945
|
unknown |
0.539 |
0.345 |
| 12 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.524 |
-2.373 |
| 13 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.522 |
-6.350 |
| 14 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.521 |
-3.549 |
| 15 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.501 |
-3.473 |
| 16 |
258209_at |
AT3G14060
|
unknown |
0.498 |
-1.532 |
| 17 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.485 |
-0.877 |
| 18 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.462 |
0.688 |
| 19 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.457 |
-2.879 |
| 20 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.441 |
-2.219 |
| 21 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.436 |
0.497 |
| 22 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.433 |
-1.205 |
| 23 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.432 |
-3.198 |
| 24 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.420 |
-1.306 |
| 25 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.419 |
-1.275 |
| 26 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.405 |
-1.303 |
| 27 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.405 |
0.009 |
| 28 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.400 |
-2.023 |
| 29 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.392 |
6.700 |
| 30 |
253814_at |
AT4G28290
|
unknown |
0.372 |
-0.417 |
| 31 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.368 |
0.793 |
| 32 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
0.366 |
-1.262 |
| 33 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.363 |
-0.614 |
| 34 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.338 |
-0.321 |
| 35 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.326 |
0.250 |
| 36 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.326 |
-0.133 |
| 37 |
259856_at |
AT1G68440
|
unknown |
0.323 |
-0.172 |
| 38 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.322 |
0.035 |
| 39 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.322 |
-0.609 |
| 40 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.320 |
-0.283 |
| 41 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.319 |
-0.732 |
| 42 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.314 |
-0.793 |
| 43 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.312 |
-0.189 |
| 44 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.311 |
-0.818 |
| 45 |
255733_at |
AT1G25400
|
unknown |
0.310 |
-0.690 |
| 46 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.309 |
-0.321 |
| 47 |
266658_at |
AT2G25735
|
unknown |
0.309 |
-1.312 |
| 48 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.305 |
-0.620 |
| 49 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.299 |
-0.385 |
| 50 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.298 |
-3.873 |
| 51 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.297 |
-2.470 |
| 52 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.296 |
-0.163 |
| 53 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.294 |
-2.369 |
| 54 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.292 |
-0.006 |
| 55 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.288 |
-1.377 |
| 56 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.287 |
-0.625 |
| 57 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.286 |
-2.657 |
| 58 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.286 |
-0.456 |
| 59 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.285 |
-1.045 |
| 60 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.283 |
0.042 |
| 61 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.277 |
-0.437 |
| 62 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.276 |
-0.329 |
| 63 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.270 |
-0.107 |
| 64 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.269 |
-2.183 |
| 65 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.268 |
3.836 |
| 66 |
252773_at |
AT3G42910
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G07240.1)] |
0.267 |
0.065 |
| 67 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.266 |
0.617 |
| 68 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.266 |
-0.258 |
| 69 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.264 |
1.218 |
| 70 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.263 |
-4.652 |
| 71 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.263 |
-0.378 |
| 72 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.261 |
-1.474 |
| 73 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.261 |
-3.446 |
| 74 |
254208_at |
AT4G24175
|
unknown |
0.260 |
0.481 |
| 75 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.259 |
-0.270 |
| 76 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.259 |
-4.074 |
| 77 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.259 |
-2.625 |
| 78 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.258 |
-2.101 |
| 79 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.257 |
1.415 |
| 80 |
263515_at |
AT2G21640
|
unknown |
0.255 |
0.702 |
| 81 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.254 |
-3.174 |
| 82 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.253 |
-0.337 |
| 83 |
262364_at |
AT1G72860
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.253 |
-0.344 |
| 84 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.253 |
-6.812 |
| 85 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
0.250 |
-1.698 |
| 86 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.249 |
-0.907 |
| 87 |
267319_at |
AT2G34660
|
ABCC2, ATP-binding cassette C2, AtABCC2, Arabidopsis thaliana ATP-binding cassette C2, ATMRP2, multidrug resistance-associated protein 2, EST4, MRP2, multidrug resistance-associated protein 2 |
0.248 |
-0.622 |
| 88 |
247913_at |
AT5G57510
|
unknown |
0.247 |
1.754 |
| 89 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.247 |
-0.576 |
| 90 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.247 |
0.020 |
| 91 |
249889_at |
AT5G22540
|
unknown |
0.246 |
2.035 |
| 92 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.244 |
-0.604 |
| 93 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
0.244 |
-0.595 |
| 94 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
0.243 |
-0.732 |
| 95 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.241 |
-1.416 |
| 96 |
266154_at |
AT2G12190
|
[Cytochrome P450 superfamily protein] |
0.241 |
0.575 |
| 97 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.239 |
1.786 |
| 98 |
260049_at |
AT1G29940
|
NRPA2, nuclear RNA polymerase A2 |
0.238 |
-0.759 |
| 99 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.236 |
-1.991 |
| 100 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
0.235 |
-0.858 |
| 101 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.235 |
-2.453 |
| 102 |
250507_at |
AT5G09940
|
unknown |
0.233 |
0.065 |
| 103 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.232 |
-1.174 |
| 104 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.231 |
-0.849 |
| 105 |
248299_at |
AT5G53080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.229 |
0.158 |
| 106 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.229 |
-0.089 |
| 107 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.228 |
-0.591 |
| 108 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
0.227 |
-1.022 |
| 109 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.227 |
1.279 |
| 110 |
264894_at |
AT1G23040
|
[hydroxyproline-rich glycoprotein family protein] |
0.225 |
0.922 |
| 111 |
265484_at |
AT2G15820
|
OTP51, ORGANELLE TRANSCRIPT PROCESSING 51 |
0.222 |
0.580 |
| 112 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.221 |
-1.012 |
| 113 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
0.221 |
-4.668 |
| 114 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.218 |
-0.865 |
| 115 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.218 |
-0.771 |
| 116 |
257866_at |
AT3G17770
|
[Dihydroxyacetone kinase] |
0.217 |
0.102 |
| 117 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.217 |
0.143 |
| 118 |
253265_at |
AT4G34040
|
MBR2, MED25 BINDING RING-H2 PROTEIN 2 |
0.217 |
0.161 |
| 119 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.215 |
0.726 |
| 120 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.214 |
-0.209 |
| 121 |
253297_at |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
0.213 |
-0.075 |
| 122 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.213 |
3.570 |
| 123 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.213 |
0.216 |
| 124 |
267459_at |
AT2G33850
|
unknown |
-1.151 |
0.579 |
| 125 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.695 |
4.748 |
| 126 |
247719_at |
AT5G59305
|
unknown |
-0.637 |
2.249 |
| 127 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.613 |
-0.684 |
| 128 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.570 |
0.744 |
| 129 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.537 |
0.358 |
| 130 |
249167_at |
AT5G42860
|
unknown |
-0.529 |
0.004 |
| 131 |
252063_at |
AT3G51590
|
LTP12, lipid transfer protein 12 |
-0.525 |
4.855 |
| 132 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.518 |
2.677 |
| 133 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.515 |
3.847 |
| 134 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.503 |
0.653 |
| 135 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.467 |
1.719 |
| 136 |
251646_at |
AT3G57780
|
unknown |
-0.456 |
-0.251 |
| 137 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.448 |
-0.154 |
| 138 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-0.444 |
5.938 |
| 139 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.441 |
1.581 |
| 140 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.440 |
1.478 |
| 141 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.421 |
5.445 |
| 142 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.420 |
2.370 |
| 143 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.404 |
2.743 |
| 144 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.403 |
3.583 |
| 145 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.397 |
2.580 |
| 146 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.392 |
-0.579 |
| 147 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.386 |
6.429 |
| 148 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
-0.386 |
4.524 |
| 149 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.385 |
5.472 |
| 150 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.383 |
0.817 |
| 151 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.381 |
1.031 |
| 152 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.372 |
5.181 |
| 153 |
255876_at |
AT2G40480
|
unknown |
-0.370 |
0.142 |
| 154 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.362 |
2.699 |
| 155 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.361 |
0.569 |
| 156 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
-0.354 |
4.557 |
| 157 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.350 |
0.065 |
| 158 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.342 |
-0.856 |
| 159 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.342 |
3.063 |
| 160 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.341 |
0.735 |
| 161 |
248509_at |
AT5G50335
|
unknown |
-0.341 |
1.490 |
| 162 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.340 |
-0.110 |
| 163 |
250689_at |
AT5G06610
|
unknown |
-0.339 |
1.065 |
| 164 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.336 |
1.230 |
| 165 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.334 |
-1.191 |
| 166 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.333 |
0.403 |
| 167 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.330 |
3.654 |
| 168 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.321 |
-0.826 |
| 169 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.318 |
-0.888 |
| 170 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.316 |
0.741 |
| 171 |
254341_at |
AT4G22130
|
SRF8, STRUBBELIG-receptor family 8 |
-0.316 |
-0.593 |
| 172 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.312 |
1.170 |
| 173 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.311 |
3.411 |
| 174 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.310 |
2.319 |
| 175 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.310 |
1.190 |
| 176 |
252361_at |
AT3G48490
|
unknown |
-0.309 |
0.565 |
| 177 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
-0.309 |
1.134 |
| 178 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.309 |
5.828 |
| 179 |
264987_at |
AT1G27030
|
unknown |
-0.308 |
-0.715 |
| 180 |
245442_at |
AT4G16710
|
[glycosyltransferase family protein 28] |
-0.307 |
0.290 |
| 181 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.307 |
2.975 |
| 182 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.305 |
0.524 |
| 183 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.302 |
0.865 |
| 184 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.299 |
0.240 |
| 185 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.298 |
1.221 |
| 186 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-0.298 |
3.082 |
| 187 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.294 |
0.470 |
| 188 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-0.293 |
0.926 |
| 189 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.291 |
1.404 |
| 190 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.290 |
0.533 |
| 191 |
256490_at |
AT1G31460
|
unknown |
-0.287 |
0.523 |
| 192 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.283 |
0.145 |
| 193 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.282 |
0.097 |
| 194 |
254362_at |
AT4G22160
|
unknown |
-0.281 |
1.343 |
| 195 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.280 |
0.246 |
| 196 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.275 |
-0.965 |
| 197 |
254193_at |
AT4G23870
|
unknown |
-0.275 |
0.495 |
| 198 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.274 |
0.168 |
| 199 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.274 |
-0.055 |
| 200 |
253772_at |
AT4G28395
|
A7, ANTHER 7, ATA7, ARABIDOPSIS THALIANA ANTHER 7 |
-0.266 |
3.757 |
| 201 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.264 |
0.208 |
| 202 |
251046_at |
AT5G02370
|
[ATP binding microtubule motor family protein] |
-0.264 |
0.435 |
| 203 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.264 |
2.107 |
| 204 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.262 |
0.683 |
| 205 |
260211_at |
AT1G74440
|
unknown |
-0.262 |
1.724 |
| 206 |
260395_at |
AT1G69780
|
ATHB13 |
-0.259 |
-0.685 |
| 207 |
263971_at |
AT2G42800
|
AtRLP29, receptor like protein 29, RLP29, receptor like protein 29 |
-0.257 |
0.597 |
| 208 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.256 |
-0.135 |
| 209 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.254 |
-1.625 |
| 210 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.253 |
-0.370 |
| 211 |
258731_at |
AT3G11880
|
unknown |
-0.253 |
0.114 |
| 212 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.248 |
-0.254 |
| 213 |
245598_at |
AT4G14200
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.247 |
0.164 |
| 214 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.247 |
1.383 |
| 215 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.243 |
-0.498 |
| 216 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.243 |
0.309 |
| 217 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.242 |
1.135 |
| 218 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.241 |
0.281 |
| 219 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.239 |
-0.781 |
| 220 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.237 |
-0.996 |
| 221 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
-0.236 |
0.134 |
| 222 |
262494_at |
AT1G21810
|
unknown |
-0.236 |
0.030 |
| 223 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.234 |
0.488 |
| 224 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.233 |
2.193 |
| 225 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.231 |
2.263 |
| 226 |
263513_at |
AT2G12400
|
unknown |
-0.231 |
-0.347 |
| 227 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.231 |
2.564 |
| 228 |
252322_at |
AT3G48550
|
unknown |
-0.229 |
1.552 |
| 229 |
258213_at |
AT3G17950
|
unknown |
-0.228 |
0.981 |
| 230 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.228 |
0.842 |
| 231 |
251788_at |
AT3G55420
|
unknown |
-0.228 |
0.649 |
| 232 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
-0.225 |
0.740 |
| 233 |
246905_at |
AT5G25570
|
unknown |
-0.221 |
0.522 |
| 234 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.220 |
1.216 |
| 235 |
263020_at |
AT1G23880
|
[NHL domain-containing protein] |
-0.219 |
0.006 |
| 236 |
250175_at |
AT5G14390
|
[alpha/beta-Hydrolases superfamily protein] |
-0.218 |
0.677 |
| 237 |
260365_at |
AT1G70630
|
RAY1, REDUCED ARABINOSE YARIV 1 |
-0.217 |
0.343 |
| 238 |
264998_at |
AT1G67330
|
unknown |
-0.217 |
-1.405 |
| 239 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
-0.217 |
0.885 |
| 240 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.216 |
2.030 |
| 241 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.216 |
-0.530 |
| 242 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
-0.215 |
0.320 |
| 243 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.215 |
0.780 |
| 244 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
-0.215 |
1.580 |
| 245 |
258196_at |
AT3G13980
|
unknown |
-0.214 |
0.064 |